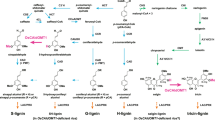
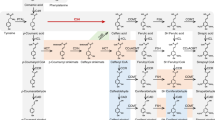
Abstract
Plant lignification exhibits notable plasticity. Lignin in many species, including Populus spp., has long been known to be decorated with p-hydroxybenzoates. However, the molecular basis for such structural modification remains undetermined. Here, we report the identification and characterization of a Populus BAHD family acyltransferase that catalyses monolignol p-hydroxybenzoylation, thus controlling the formation of p-hydroxybenzoylated lignin structures. We reveal that Populus acyltransferase PHBMT1 kinetically preferentially uses p-hydroxybenzoyl-CoA to acylate syringyl lignin monomer sinapyl alcohol in vitro. Consistently, disrupting PHBMT1 in Populus via CRISPR–Cas9 gene editing nearly completely depletes p-hydroxybenzoates of stem lignin; conversely, overexpression of PHBMT1 enhances stem lignin p-hydroxybenzoylation, suggesting PHBMT1 functions as a prime monolignol p-hydroxybenzoyltransferase in planta. Altering lignin p-hydroxybenzoylation substantially changes the lignin solvent dissolution rate, indicative of its structural significance on lignin physiochemical properties. Identification of monolignol p-hydroxybenzoyltransferase offers a valuable tool for tailoring lignin structure and physiochemical properties and for engineering the industrially important platform chemical in woody biomass.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the paper and/or its Supplementary Information. Other publicly available datasets: BAHD family proteins, NCBI (https://www.ncbi.nlm.nih.gov/); the PtrPHBMT1 homologues in Populus, Phytozome V12 (https://phytozome.jgi.doe.gov/pz/portal.html); the gene chip dataset, PlaNet (http://www.gene2function.de); gene co-expression data; Phytozome v.12.1 (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ptrichocarpa).
References
Boerjan, W., Ralph, J. & Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 (2003).
Ragauskas, A. J. et al. Lignin valorization: improving lignin processing in the biorefinery. Science 344, 1246843 (2014).
Ralph, J. Hydroxycinnamates in lignification. Phytochem. Rev. 9, 65–83 (2010).
Ralph, J. et al. Peroxidase dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochem. Rev. 3, 79–96 (2004).
Del Rio, J. C. et al. Highly acylated (acetylated and/or p-coumaroylated) native lignins from diverse herbaceous plants. J. Agric. Food Chem. 56, 9525–9534 (2008).
Del Rio, J. C. et al. Structural characterization of the lignin in the cortex and pith of elephant grass (Pennisetum purpureum) stems. J. Agric. Food Chem. 60, 3619–3634 (2012).
Karlen, S. D. et al. Commelinid monocotyledon lignins are acylated by p-coumarate. Plant Physiol. 177, 513–521 (2018).
Karlen, S. D. et al. Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci. Adv. 2, e1600393 (2016).
Smith, D. C. C. para-Hydroxybenzoate groups in the lignin of aspen (Populus tremula). J. Chem. Soc. 3, 2347–2351 (1955).
Morreel, K. et al. Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol. 136, 3537–3549 (2004).
Lu, F., Ralph, J., Morreel, K., Messens, E. & Boerjan, W. Preparation and relevance of a cross-coupling product between sinapyl alcohol and sinapyl p-hydroxybenzoate. Org. Biomol. Chem. 2, 2888–2890 (2004).
Landucci, L. L., Deka, G. C. & Roy, D. N. A C13 NMR study of milled wood lignins from hybrid salix clones. Holzforschung 46, 505–511 (1992).
Lu, F. C. et al. Naturally p-hydroxybenzoylated lignins in palms. Bioenerg. Res. 8, 934–952 (2015).
Del Rio, J. C., Marques, G., Rencoret, J., Martinez, A. T. & Gutierrez, A. Occurrence of naturally acetylated lignin units. J. Agric. Food Chem. 55, 5461–5468 (2007).
Hatfield, R. D. et al. Grass lignin acylation: p-coumaroyl transferase activity and cell wall characteristics of C3 and C4 grasses. Planta 229, 1253–1267 (2009).
Withers, S. et al. Identification of grass-specific enzyme that acylates monolignols with p-coumarate. J. Biol. Chem. 287, 8347–8355 (2012).
Petrik, D. L. et al. p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J. 77, 713–726 (2014).
Marita, J. M., Hatfield, R. D., Rancour, D. M. & Frost, K. E. Identification and suppression of the p-coumaroyl CoA:hydroxycinnamyl alcohol transferase in Zea mays L. Plant J. 78, 850–864 (2014).
Wilkerson, C. G. et al. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344, 90–93 (2014).
St-Pierre, B. & De Luca, V. in Recent Advances in Phytochemistry. Evolution of Metabolic Pathways Vol. 34 (eds Romeo, J. T. et al.) 285–315 (Elsevier, 2000).
D’Auria, J. C. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9, 331–340 (2006).
Sannigrahi, P., Ragauskas, A. J. & Tuskan, G. A. Poplar as a feedstock for biofuels: a review of compositional characteristics. Biofuels Bioprod. Bioref. 4, 209–226 (2010).
Andersson-Gunneras, S. et al. Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 45, 144–165 (2006).
Kim, H. et al. Monolignol benzoates incorporate into the lignin of transgenic Populus trichocarpa depleted in C3H and C4H. ACS Sustain. Chem. Eng. 8, 3644–3654 (2020).
Tuskan, G. A. E. A. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Yu, X.-H., Gou, J.-Y. & Liu, C. J. BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: bioinformatics and gene expression. Plant Mol. Biol. 70, 421–442 (2009).
Ralph, J. & Lu, F. The DFRC method for lignin analysis. 6. A simple modification for identifying natural acetates on lignins. J. Agric. Food Chem. 46, 4616–4619 (1998).
Boatright, J. et al. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135, 1993–2011 (2004).
Chedgy, R. J., Kollner, T. G. & Constabel, C. P. Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry 113, 149–159 (2015).
Dexter, R. et al. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 49, 265–275 (2007).
Daniel, B. et al. Oxidation of monolignols by members of the berberine bridge enzyme family suggests a role in plant cell wall metabolism. J. Biol. Chem. 290, 18770–18781 (2015).
Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S. & Yang, S. H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264 (2015).
Cai, Y. et al. Enhancing digestibility and ethanol yield of Populus wood via expression of an engineered monolignol 4-O-methyltransferase. Nat. Commun. 7, 11989 (2016).
Lu, F. & Ralph, J. Detection and determination of p-coumaroylated units in lignins. J. Agric. Food Chem. 47, 1988–1992 (1999).
Mansfield, S. D., Kim, H., Lu, F. & Ralph, J. Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7, 1579–1589 (2012).
Kim, H. & Ralph, J. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org. Biomol. Chem. 8, 576–591 (2010).
Regner, M., Bartuce, A., Padmakshan, D., Ralph, J. & Karlen, S. D. Reductive cleavage method for quantitation of monolignols and low-qbundance monolignol conjugates. ChemSusChem 11, 1600–1605 (2018).
Meyermans, H. et al. Modifications in lignin and accumulation of phenolic glycosides in poplar xylem upon down-regulation of caffeoyl coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275, 36899–36909 (2000).
Henry, R. A., Kuo, Y. M., Bhattacharjee, V., Yen, T. J. & Andrews, A. J. Changing the selectivity of p300 by acetyl-CoA modulation of histone acetylation. ACS Chem. Biol. 10, 146–156 (2015).
Denisov, I. G. & Sligar, S. G. A novel type of allosteric regulation: functional cooperativity in monomeric proteins. Arch. Biochem. Biophys. 519, 91–102 (2012).
Sibout, R. et al. Structural redesigning Arabidopsis lignins into alkali-soluble lignins through the expression of p-coumaroyl-CoA:monolignol transferase PMT. Plant Physiol. 170, 1358–1366 (2016).
Smith, R. A. et al. Engineering monolignol p-coumarate conjugates into poplar and Arabidopsis lignins. Plant Physiol. 169, 2992–3001 (2015).
Wang, S., Bilal, M., Hu, H., Wang, W. & Zhang, X. 4-Hydroxybenzoic acid—a versatile platform intermediate for value-added compounds. Appl. Microbiol. Biotechnol. 102, 3561–3571 (2018).
Yang, M. et al. Accumulation of high-value bioproducts in planta can improve the economics of advanced biofuels. Proc. Natl Acad. Sci. USA 117, 8639–8648 (2020).
Biegert, T., Altenschmidt, U., Eckerskorn, C. & Fuchs, G. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur. J. Biochem. 213, 555–561 (1993).
Stockigt, J. & Zenk, M. H. Chemical syntheses and properties of hydroxycinnamoyl-coenzyme A derivatives. Z. Naturforsch. 30C, 352–358 (1975).
Yu, X.-H. & Liu, C.-J. Development of an analytical method for genome-wide functional identification of plant acyl-CoA dependent acyltransferases. Anal. Biochem. 358, 146–148 (2006).
Gou, M. et al. Cytochrome b5 is an obligate electron shuttle protein for syringyl lignin biosynthesis in Arabidopsis. Plant Cell 31, 1344–1366 (2019).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Liu, H. et al. CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532 (2017).
Mao, Y. et al. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011 (2013).
Ma, C., Strauss, S. F. & Meilan, R. Agrobacterium-mediated transformation of the genome-sequenced poplar clone, Nisqually-1 (Populus trichocarpa). Plant Mol. Biol. Rep. 22, 1–9 (2004).
Ma, X., Chen, L., Zhu, Q., Chen, Y. & Liu, Y. G. Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant 8, 1285–1287 (2015).
Printz, B. et al. An improved protocol to study the plant cell wall proteome. Front. Plant Sci. 6, 237 (2015).
Foster, C. E., Martin, T. M. & Pauly, M. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass). Part I: lignin. J. Vis. Exp. 11, 1745 (2010).
Karlen, S. D. et al. Highly decorated lignins in leaf tissues of the canary island date palm Phoenix canariensis. Plant Physiol. 175, 1058–1067 (2017).
Tobimatsu, Y. et al. Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25, 2587–2600 (2013).
Lam, P. Y. et al. Recruitment of specific flavonoid B-ring hydroxylases for two independent biosynthesis pathways of flavone-derived metabolites in grasses. New Phytol. 223, 204–219 (2019).
Acknowledgements
This work was initiated with the support of United States Department of Energy (DOE)-USDA joint Plant Feedstock Genomics Program (FWP no. Bo-135, 2006) and the Laboratory Directed Research and Development Program (LDRD-07-047) of Brookhaven National Laboratory (to C.J.L) for poplar BAHD enzyme functional screening. Generation of poplar knockout lines and chemical analyses were partly supported by the Joint BioEnergy Institute, one of the Bioenergy Research Centers of the US DOE, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US DOE. Enzyme characterization, transgenic poplar generation and chemical analysis were also partially supported by the DOE, Office of Science, Office of Basic Energy Sciences, specifically the Physical Biosciences program of the Chemical Sciences, Geosciences and Biosciences Division under contract no. DE-SC0012704 (to C.-J.L.). P.-Y.L. and Y.T. acknowledge research grants from the Japan Society for the Promotion of Science (grant no. JP20H03044) for NMR analysis and a part of the study was conducted using the facilities in the DASH/FBAS of RISH, Kyoto University and the NMR spectrometer at the JURC of ICR, Kyoto University.
Author information
Authors and Affiliations
Contributions
C.-J.L., Y.Z. and X.Y. conceived the research plan and designed the experiments. X.Y. cloned the genes and conducted initial functional screening. Y.Z. conducted enzymatic kinetic analysis, designed CRISPR–Cas9 gene editing strategy and generated and analysed knockout transgenic lines. K.Z. generated overexpression lines. Y.T. and P-Y.L. conducted NMR analysis. C.-J.L., Y.Z., X.Y. and Y.T. analysed and interpreted data. C.-J.L., Y.Z. and X.Y. wrote the manuscript; all the authors edited the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Gerald Tuskan, Yihua Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Preliminary enzymatic activity screening of the recombinant BAHD acyltransferase candidates from P. trichocarpa.
Schema of the postulated monolignol acylation reactions in Populus and preliminary screening of enzymatic activities of the recombinant BAHD acyltransferase candidates from P. trichocarpa in catalysing conjugation of sinapyl alcohol with acetyl-CoA. Only PtACT54 exhibited a detectable activity when the enzymes were incubated with both substrates. The data are presented as means ± S. D. of three independent experiments.
Extended Data Fig. 2 The apparent optimal temperature and pH value of buffer system of PtrPHBMT1(PtACT54)-catalysed reactions.
a, b, The pH preference of PtrPHBMT1(PtACT54) with p-hydroxybenzoyl-CoA (a) or acetyl-CoA (b) and sinapyl alcohol substates. c, d, The optimal temperature for PtrPHBMT1(PtACT54)-catalysed reaction with p-hydroxybenzoyl-CoA (c) or acetyl-CoA (d) and sinapyl alcohol substrates. The data in (a–d) are presented as means ± S. D. of three (b) or four (a, c, d) experimental repeats. In (a, b) the measured activity at pH 5 was set as 100%. In (c and d) the activity at 20 °C was set as 100%.
Extended Data Fig. 3 Kinetic plots of PtrPHBMT1 against thioester donor substrates.
a, A plot of enzymatic activity as a function of p-hydroxybenzoyl-CoA concentration that obeys Michaelis–Menten kinetics. b, A plot of enzymatic activity as a function of benzoyl-CoA concentration that obeys Hill kinetics. c, A plot of enzymatic activity as a function of acetyl-CoA concentration that obeys Hill kinetics. The reactions were conducted at the fixed concentration of sinapyl alcohol with varied concentrations of thioesters in MES buffer (pH 5.5) at 25 °C for 15 min. Each data point represents mean of at least three replicates.
Extended Data Fig. 4 Expression pattern of PtrPHBMT1 in Populus.
a, Relative expression levels of PtrPHBMT1 was determined via RT–qPCR using RNAs from P. trichocarpa leaf, root, stem (> 1 year), shoot apex, and the bark from the young stem (<3 months), the developing xylem scrapped from young stem (<3 months), the debarked young (<3 months) and old wood (>1 year). Data are presented as means ± S. D. of three biological repeats. The relative expression levels were referenced with ubiquitin gene. The expression level in the old wood sample was set as 100%. b, In silico gene expression pattern of PtrPHBMT1. The data were retrieved from PlaNet (http://aranet.sbs.ntu.edu.sg/) using Probset ID PtpAffx.59062.1.A1_at. Data are presented as means ± S. D. of signal values from three individual microarray experiments.
Extended Data Fig. 5 Subcellular localization of PtrPHBMT1.
a, b, Fluorescence distribution of the free GFP(a) and PtrPHBMT1–GFP fusion protein (b) transiently expressed in N. benthamiana leaf cells. c, d, Plasmolysis analysis for GFP (c) and PtrPHBMT1–GFP (d) transiently expressed in N. benthamiana leaf cells. The GFP fluorescence in green and chlorophyll fluorescence in red. Bar = 20 μm. Arrows point out the cell wall region. e, Immunoblots of PHBMT1 in the soluble (S), microsomal (MS), and cell (CW) protein fractions using anti-PHBMT1 antibody. 20 µg proteins were loaded in each lane. M, marker. The experiments in (a–e) were repeated twice independently with similar results.
Extended Data Fig. 6 DNA sequences of PHBMT1 target site and the potential off-target site of its close homologous gene in the obtained CRISPR/Cas9 transgenic lines.
The homozygous, biallelic, and chimeric mutations were determined according to the allelic sequences of the target site compared to Populus PHBMT1 reference sequence. PAM sequence(AGG) is highlighted. The edited events usually happen after -3 position of PAM (g1-8, g1-9, g1-11). The mutations are occurred in the targeted PtxaPHBMT1 gene but not in its homologue Ptxa001G447300.
Extended Data Fig. 7 Analysis of CRISPR/Cas9-mediated PHBMT1 knockout hybrid aspens.
a, Genotyping of the transgenic lines with selection marker NPT II gene. The experiment was repeated twice independently with the same results. Marker,Thermo Scientific GeneRuler 1 kb DNA ladder. b, Immunoblots with anti-PHBMT1 antibody on the crude proteins from the WT, non-edited control and PHBMT1 knockout transgenic lines. The immunoblots against anti-actin antibody served as the loading control. The original blots are presented in Supplemental Fig. 5a and c. This experiment was repeated twice independently with similar results. c, Growth phenotype of 2-month-old hybrid aspens. Bar = 4 cm. d, Growth phenotype of 6-month-old plants. Bar = 30 cm. e, The basal stem (30 cm) of the 6-month-old WT, non-edited control, and PHBMT1 knockout lines. f, Total acetyl bromide soluble lignin content in the stem of 2-month-old plantlets. Data are presented as means ± S. D. of three biological replicates. Asterisk indicates significant difference compared to the WT, * P < 0.05 (Student’s t-test, with two-tailed distribution, two-sample unequal variance). P = 3.76×10−4 (g1-8), 3.03×10−4 (g1-9), and 0.03 (g1-10). CWR, cell wall residues. g, Stem height of 3- and 5-month-old plants. h, Basal stem diameter of 3- and 5-month-old plants. i, Woody biomass yield of 6-month-old hybrid aspens. j, The calculated wood density of the basal stems of 6-month-old plants. k, l, Total acetyl bromide lignin content (k) and monomeric composition (l) of 6-month-old basal stems. CWR, cell wall residues. G, Guaiacyl monomer. S, Syringyl monomer. The data in (g–l) are presented as means ± S. D. of three biological replicates for WT, Ctrl, g1-9, g1-11 and five biological replicates for g1-8. No statistical differences were found between the knockout and the WT and/or non-edited control (Ctrl) plants.
Extended Data Fig. 8 Analysis of PHBMT1 overexpression hybrid aspens.
a, Growth phenotype of 6-month-old PtrPHBMT1 overexpression aspens (OEs). Bar = 30 cm. b, Immunoblots probed with anti-PHBMT1 antibody on the crude proteins from the WT and PHBMT1 OE transgenic aspens. The immunoblots against anti-actin antibody served as the loading control. The original blots are presented in Supplementary Fig. 5b and d. The experiments were repeated twice independently with similar results. c, d, Stem height (c) and basal stem diameter (d) of the 3- and 5-month-old WT and OE lines. The data in (c, d) are presented as means ± S. D. of four (WT and OE3) and seven (OE1) biological repeats. e, Total acetyl bromide soluble lignin content of 6-month-old stems. The data are presented as means ± S. D. of three biological repeats. f, Woody biomass yield of the 6-month-old hybrid aspens. g, The calculated wood density of the basal stems of 6-month-old plants. The data in (f, g) are presented as means ± S. D. of three (WT and OE3) and four (OE1) biological repeats. No statistical differences were found relative to the WT.
Extended Data Fig. 9 Detection of sinapyl p-hydroxybenzoate (SA-pBA) from poplar lignin fraction via DFRC.
a–c, Extract ion chromatography of the DFRC products from the prepared cellulolytic enzyme lignin of the PHBMT1 overexpression line OE1(a), the WT (b) and the PHBMT1 knockout mutant g1-8 (c), The ions were scanned at m/z = 372 for 4-O-acetylsinapyl-p-O-acetylbenzoate. Note the different ion abundance scale in a–c. d, The full mass spectra of the peak at retention time 39 min in a, b which is absent in c. e, Schematic diagram of 4-O-acetylsinapyl-p-O-acetylbenzoate, the DFRC product of SA-pBA, and its characteristic ions.
Extended Data Fig. 10 Quantification of wall-bound acetate in the poplar WT and transgenic lines.
a, Acetate (AA) content released from the cell wall residues (CWR) of the 2-month-old WT, non-edited control (Ctrl) and PHBMT1 knockout lines. The data are presented as means ± S. D. of four biological repeats. b, Acetate content released from the CWRs of 6-month-old WT, non-edited control and PHBMT1 knockout lines. The data are presented as means ± S. D. of three (WT, Ctrl, g1-9, g1-11) or five (g1-8) biological replicates. c, Acetate content released from the CWRs of 6-month-old WT and PHBMT1 overexpression (OE) lines. The data are presented as means ± S. D. of three biological repeats. d, Acetate content released from the prepared cellulolytic enzyme lignin (CEL) of the 6-month-old WT, PHBMT1 knockout (phbmt1, combination of g1-8, g1-9 and g1-11) and overexpression line (OE1). The data are presented as means ± S. D. of three biological replicates. Asterisk indicates significant difference compared to the WT with P = 0.012 (Student’s t-test, with two-tailed distribution, two-sample unequal variance).
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Tables 1–3.
Rights and permissions
About this article
Cite this article
Zhao, Y., Yu, X., Lam, PY. et al. Monolignol acyltransferase for lignin p-hydroxybenzoylation in Populus. Nat. Plants 7, 1288–1300 (2021). https://doi.org/10.1038/s41477-021-00975-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00975-1