Abstract
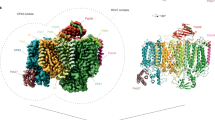
Photosystem II (PSII) is a multisubunit pigment–protein complex and catalyses light-induced water oxidation, leading to the conversion of light energy into chemical energy and the release of dioxygen. We analysed the structures of two Psb28-bound PSII intermediates, Psb28–RC47 and Psb28–PSII, purified from a psbV-deletion strain of the thermophilic cyanobacterium Thermosynechococcus vulcanus, using cryo-electron microscopy. Both Psb28–RC47 and Psb28–PSII bind one Psb28, one Tsl0063 and an unknown subunit. Psb28 is located at the cytoplasmic surface of PSII and interacts with D1, D2 and CP47, whereas Tsl0063 is a transmembrane subunit and binds at the side of CP47/PsbH. Substantial structural perturbations are observed at the acceptor side, which result in conformational changes of the quinone (QB) and non-haem iron binding sites and thus may protect PSII from photodamage during assembly. These results provide a solid structural basis for understanding the assembly process of native PSII.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The cryo-EM density maps and atomic models of the Psb28–RC47 and Psb28–PSII complexes at 3.14 Å resolution have been deposited in the Electron Microscopy Data Bank and the Protein Data Bank (EMD ID 30902 and PDB ID 7DXA for Psb28–RC47, EMD ID 30909 and PDB ID 7DXH for Psb28–PSII). The data that support the findings of this study are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Barber, J. Photosystem II: the engine of life. Q. Rev. Biophys. 36, 71–89 (2003).
Shen, J.-R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant. Biol. 66, 23–48 (2015).
Umena, Y., Kawakami, K., Shen, J. R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011).
Suga, M. et al. Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517, 99–103 (2015).
Ago, H. et al. Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 291, 5676–5687 (2016).
Wei, X. et al. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 534, 69–74 (2016).
Pi, X. et al. The pigment–protein network of a diatom photosystem II–light-harvesting antenna supercomplex. Science 365, eaax4406 (2019).
Nagao, R. et al. Structural basis for energy harvesting and dissipation in a diatom PSII–FCPII supercomplex. Nat. Plants 5, 890–901 (2019).
Shen, L. et al. Structure of a C2S2M2N2-type PSII–LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 116, 21246–21255 (2019).
Sheng, X. et al. Structural insight into light harvesting for photosystem II in green algae. Nat. Plants 5, 1320–1330 (2019).
Nixon, P. J., Michoux, F., Yu, J., Boehm, M. & Komenda, J. Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16 (2010).
Boehm, M. et al. Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: isolation of CP47 and CP43 complexes. J. Biol. Chem. 286, 14812–14819 (2011).
Boehm, M. et al. Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: implications for the assembly and repair of photosystem II. Phil. Trans. R. Soc. B 367, 3444–3454 (2012).
Komenda, J., Sobotka, R. & Nixon, P. J. Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251 (2012).
Zhang, M. et al. Structural insights into the light-driven auto-assembly process of the water-oxidizing Mn4CaO5-cluster in photosystem II. eLife 6, e26933 (2017).
Gisriel, C. J. et al. Cryo-EM structure of monomeric photosystem II from Synechocystis sp. PCC 6803 lacking the water-oxidation complex. Joule 4, 2131–2148 (2020).
Tokano, T., Kato, Y., Sugiyama, S., Uchihashi, T. & Noguchi, T. Structural dynamics of a protein domain relevant to the water-oxidizing complex in photosystem II as visualized by high-speed atomic force microscopy. J. Phys. Chem. B 124, 5847–5857 (2020).
Huang, G. et al. Structural insights into a dimeric Psb27–photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus. Proc. Natl Acad. Sci. USA 118, e2018053118 (2021).
Zabret, J. et al. Structural insights into photosystem II assembly. Nat. Plants 7, 524–538 (2021).
Bao, H. & Burnap, R. L. Photoactivation: the light-driven assembly of the water oxidation complex of photosystem II. Front. Plant Sci. 7, 578 (2016).
Nickelsen, J. & Rengstl, B. Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635 (2013).
Jarvi, S., Suorsa, M. & Aro, M. E. Photosystem II repair in plant chloroplasts—regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847, 900–909 (2015).
Heinz, S., Liauw, P., Nickelsen, J. & Nowaczyk, M. Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857, 274–287 (2016).
Kashino, Y. et al. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 (2002).
Nowaczyk, M. M. et al. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18, 3121–3131 (2006).
Avramov, A. P., Hwang, H. J. & Burnap, R. L. The role of Ca2+ and protein scaffolding in the formation of nature’s water oxidizing complex. Proc. Natl Acad. Sci. USA 117, 28036–28045 (2020).
Mabbitt, P. D., Wilbanks, S. M. & Eaton-Rye, J. J. Structure and function of the hydrophilic Photosystem II assembly proteins: Psb27, Psb28 and Ycf48. Plant Physiol. Biochem. 81, 96–107 (2014).
Dobakova, M., Sobotka, R., Tichy, M. & Komenda, J. Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149, 1076–1086 (2009).
Sakata, S., Mizusawa, N., Kubota-Kawai, H., Sakurai, I. & Wada, H. Psb28 is involved in recovery of photosystem II at high temperature in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1827, 50–59 (2013).
Beckova, M. et al. Association of Psb28 and Psb27 proteins with PSII–PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol. Plant 10, 62–72 (2017).
Jung, K. H. et al. Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet. 4, e1000164 (2008).
Yao, D. et al. Localization of the small CAB-like proteins in photosystem II. J. Biol. Chem. 282, 267–276 (2007).
Liu, H., Roose, J. L., Cameron, J. C. & Pakrasi, H. B. A genetically tagged Psb27 protein allows purification of two consecutive photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J. Biol. Chem. 286, 24865–24871 (2011).
Nowaczyk, M. M. et al. Deletion of psbJ leads to accumulation of Psb27–Psb28 photosystem II complexes in Thermosynechococcus elongatus. Biochim. Biophys. Acta 1817, 1339–1345 (2012).
Weisz, D. A. et al. Mass spectrometry-based cross-linking study shows that the Psb28 protein binds to cytochrome b559 in Photosystem II. Proc. Natl Acad. Sci. USA 114, 2224–2229 (2017).
Weisz, D. A. et al. A novel chlorophyll protein complex in the repair cycle of photosystem II. Proc. Natl Acad. Sci. USA 116, 21907–21913 (2019).
Funk, C. & Vermaas, W. A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38, 9397–9404 (1999).
He, Q., Dolganov, N., Bjorkman, O. & Grossman, A. R. The high light-inducible polypeptides in Synechocystis PCC6803: expression and function in high light. J. Biol. Chem. 276, 306–314 (2001).
Komenda, J. & Sobotka, R. Cyanobacterial high-light-inducible proteins—protectors of chlorophyll-protein synthesis and assembly. Biochim. Biophys. Acta 1857, 288–295 (2016).
Yang, Y. et al. Solution NMR structure of photosystem II reaction center protein Psb28 from Synechocystis sp. strain PCC 6803. Proteins 79, 340–344 (2011).
Bialek, W. et al. Crystal structure of the Psb28 accessory factor of Thermosynechococcus elongatus photosystem II at 2.3 Å. Photosynth. Res. 117, 375–383 (2013).
Xiao, Y. et al. Role of PsbV-Tyr137 in photosystem II studied by site-directed mutagenesis in the thermophilic cyanobacterium Thermosynechococcus vulcanus. Photosynth. Res. 146, 41–54 (2020).
Pascual-Aznar, G. et al. Psb35 protein stabilizes the CP47 assembly module and associated high-light inducible proteins during the biogenesis of photosystem II in the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 61, 178–190 (2021).
Xu, H., Vavilin, D., Funk, C. & Vermaas, W. Small Cab-like proteins regulating tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 49, 149–160 (2002).
Hernandez-Prieto, M. A. et al. The small CAB-like proteins of the cyanobacterium Synechocystis sp. PCC 6803: their involvement in chlorophyll biogenesis for Photosystem II. Biochim. Biophys. Acta 1807, 1143–1151 (2011).
Xu, H., Vavilin, D., Funk, C. & Vermaas, W. Multiple deletions of small Cab-like proteins in the cyanobacterium Synechocystis sp. PCC 6803: consequences for pigment biosynthesis and accumulation. J. Biol. Chem. 279, 27971–27979 (2004).
Knoppová, J. et al. Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26, 1200–1212 (2014).
Staleva, H. et al. Mechanism of photoprotection in the cyanobacterial ancestor of plant antenna proteins. Nat. Chem. Biol. 11, 287–291 (2015).
Vavilin, D., Yao, D. & Vermaas, W. Small Cab-like proteins retard degradation of photosystem II-associated chlorophyll in Synechocystis sp. PCC 6803: kinetic analysis of pigment labeling with 15N and 13C. J. Biol. Chem. 282, 37660–37668 (2007).
Havaux, M., Guedeney, G., He, Q. F. & Grossman, A. R. Elimination of high-light-inducible polypeptides related to eukaryotic chlorophyll a/b-binding proteins results in aberrant photoacclimation in Synechocystis PCC6803. Biochim. Biophys. Acta 1557, 21–33 (2003).
Sinha, R. K., Komenda, J., Knoppova, J., Sedlarova, M. & Pospisil, P. Small CAB-like proteins prevent formation of singlet oxygen in the damaged photosystem II complex of the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Environ. 35, 806–818 (2012).
Tibiletti, T., Rehman, A. U., Vass, I. & Funk, C. The stress-induced SCP/HLIP family of small light harvesting-like proteins (ScpABCDE) protects Photosystem II from photoinhibitory damages in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 135, 103–114 (2018).
Brinkert, K., De Causmaecker, S., Krieger-Liszkay, A., Fantuzzi, A. & Rutherford, A. W. Bicarbonate-induced redox tuning in Photosystem II for regulation and protection. Proc. Natl Acad. Sci. USA 113, 12144–12149 (2016).
Liu, H., Huang, R. Y., Chen, J., Gross, M. L. & Pakrasi, H. B. Psb27, a transiently associated protein, binds to the chlorophyll binding protein CP43 in photosystem II assembly intermediates. Proc. Natl Acad. Sci. USA 108, 18536–18541 (2011).
Liu, H. et al. Mass spectrometry-based footprinting reveals structural dynamics of loop E of the chlorophyll-binding protein CP43 during photosystem II assembly in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 288, 14212–14220 (2013).
Muhlenhoff, U. & Chauvat, F. Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol. Gen. Genet. 252, 93–100 (1996).
Onai, K., Morishita, M., Kaneko, T., Tabata, S. & Ishiura, M. Natural transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: a simple and efficient method for gene transfer. Mol. Genet. Genomics 271, 50–59 (2004).
Shen, J. R., Kawakami, K. & Koike, H. Purification and crystallization of oxygen-evolving photosystem II core complex from thermophilic cyanobacteria. Methods Mol. Biol. 684, 41–51 (2011).
Sugiura, M. & Inoue, Y. Highly purified thermo-stable oxygen-evolving photosystem II core complex from the thermophilic cyanobacterium Synechococcus elongatus having His-tagged CP43. Plant Cell Physiol. 40, 1219–1231 (1999).
Ikeuchi, M. & Inoue, Y. A new 4.8-kDa polypeptide intrinsic to the PS II reaction center, as revealed by modified SDS–PAGE with improved resolution of low-molecular-weight proteins. Plant Cell Physiol. 29, 1233–1239 (1988).
Kawakami, K., Iwai, M., Ikeuchi, M., Kamiya, N. & Shen, J.-R. Location of PsbY in oxygen-evolving photosystem II revealed by mutagenesis and X-ray crystallography. FEBS Lett. 581, 4983–4987 (2007).
Schägger, H. Tricine-SDS–PAGE. Nat. Protoc. 1, 16–22 (2006).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Acknowledgements
We thank J. Lei and the staff at the Tsinghua University Branch of the National Center for Protein Sciences Beijing for providing facility support, the Explorer 100 cluster system of the Tsinghua National Laboratory for Information Science and Technology for providing computation resources, and H. Deng and X. Meng in the Proteinomics Facility at the Technology Center for Protein Sciences, Tsinghua University, for protein MS analysis. This work was supported by the National Key R&D Program of China (grant nos. 2017YFA0503700, 2016YFA0501101, 2017YFA0504600, 2020YFA0907600 and 2019YFA0906300), the National Natural Science Foundation of China (grant no. 31470339), the Strategic Priority Research Program of CAS (grant nos. XDA27050402 and XDB17000000), a CAS Key Research programme for Frontier Science (grant no. QYZDY-SSW-SMC003), Youth Innovation Promotion Association of CAS (grant no. 2020081) and CAS Interdisciplinary Innovation Team (grant no. JCTD-2020-06).
Author information
Authors and Affiliations
Contributions
G. Han, J.-R.S. and S.-F.S. conceived the project. Y.X., G. Han and Q.Z. performed the sample isolation and characterization. G. Huang took the cryo-EM images, processed the cryo-EM data and built the structure model. Y.X., G. Huang, X.Y., W.W., G. Han, S.-F.S. and J.-R.S. analysed the structure. Y.X., G. Huang, G. Han, J.-R.S. and S.-F.S. jointly wrote the manuscript, and all the authors contributed to the discussions of the results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Rob Burnap, Nathan Nelson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Purification and characterization of Psb28-PSII and Psb28-RC47 complexes from the HisPsb28-ΔPsbV and HisPsb28-WT strain of T. vulcanus.
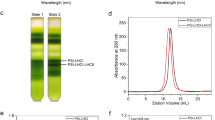
a Elution pattern of the Psb28-PSII and Psb28-RC47 complexes from the HisPsb28-ΔPsbV strain by Ni affinity chromatography column. b SDS-PAGE analysis of the purified samples shown in panel a. Lane 1: fraction I; lane 2: fraction II; lane 3: fraction III; lane M: Molecular weight marker. The band of Psb28 was indicated by a red star. Western-blotting analysis of His-Psb28 was shown in the bottom of the SDS-PAGE. c BN-PAGE analysis of the purified samples after the Ni affinity chromatography in panel a. Lane 1: fraction I; lane 2: fraction II; lane 3: fraction III. d Western-blotting analysis of native PSII and fraction III of panel a using the antibody against D1, Tsl0063 and PsbJ subunits. Lane 1: native PSII; lane 2: fraction III of panel a; lane M: Molecular weight marker. e Elution pattern of the Psb28-PSII and Psb28-RC47 complexes from the HisPsb28-WT strain by Ni affinity column. f BN-PAGE analysis of the purified samples shown in panel e. Lane 1: fraction I; lane 2: fraction II; lane 3: fraction III; lane 4: native PSII dimer with some contaminations of the PSII monomer. g Two-dimensional SDS-PAGE analysis of band 1 to band 4 shown in panel c and f. Lanes 1-2: band 1-2 from lane 3 of panel c; lanes 3-4: band 3-4 from lane 3 of panel f; lane M: Molecular weight marker. Western-blotting analyses with the antibodies against D1, His-tag and Psb27 were shown in the bottom of the SDS-PAGE. The primary antibody against D1(AS05084) was purchased from Agrisera. The primary antibody against His-tag (A02051) was purchased from Abbkine. The primary antibody against PsbJ and Tsl0063 were custom-made by Genscript, respectively. The horseradish peroxidase (HRP)-conjugated secondary antibody (Goat Anti Rabbit IgG) (AS09602) was purchased from Agrisera. Data shown in this figure is repeated more than three times, and all resulted in the same results.
Extended Data Fig. 2 Further purification and characterization of Psb28-PSII and Psb28-RC47 complexes from the HisPsb28-ΔPsbV strain of T. vulcanus.
a Elution pattern of the Psb28-PSII and Psb28-RC47 complexes (fraction III in Extended Data Fig. 1a) from a Mono Q anion-exchange column. b SDS-PAGE analysis of the purified samples shown in panel a. Lane 1: fraction 1; lane 2: fraction 2; lane 3: fraction 3; lane 4: fraction 4; lane M: Molecular weight marker. c BN-PAGE analysis of the purified samples after the Mono Q anion-exchange column. Lane 1: fraction 1; lane 2: fraction 2; lane 3: fraction 3; lane 4: fraction 4. Data shown in this figure is conducted more than three times, and all resulted in the same results.
Extended Data Fig. 3 Cryo-EM densities and structural models of the PSII core intrinsic, extrinsic subunit and various cofactors of the Psb28-RC47 complex (a) and Psb28-PSII complex (b).
The intrinsic subunits and extrinsic subunit are shown as mixed cartoon/stick model. All cofactors are shown as stick models. Fe and Mg are shown as magenta and green spheres respectively. The cryo-EM density map of each subunit is depicted in gray meshes.
Extended Data Fig. 4 Sequence comparison of Tsl0063 with other subunits that have a sequence similarity above 90%.
V5V6Y3: CAB/ELIP/HLIP family protein from Thermosynechococcus sp. NK55a; A0A3M2FSJ2: light-harvesting-like protein (Ssl1498 family) from Cyanobacteria bacterium J003; Q8DMP8: Tsl0063 protein from Thermosynechococcus elongatus strain BP-1; A0A5C2M567: light-harvesting-like protein (Ssl1498 family) from Thermosynechococcus sp. CL-1; A0A3B7MFR6: light-harvesting-like protein (Ssl1498 family) from Thermosynechococcus elongatus PKUAC-SCTE542. (These information is obtained by searching the database of https://www.uniprot.org/).
Extended Data Fig. 5 Structural comparison of the Psb28-RC47 and Psb28-PSII complexes of T. vulcanus.
a, b Structural comparison of the Psb28-RC47 and Psb28-PSII complexes of T. vulcanus, viewed along the membrane plane (a) and from the stromal side (b). All subunits of Psb28-RC47 are shown in grey and the subunits of Psb28-PSII are colored differently as those in Fig. 1.
Extended Data Fig. 6 Structural comparison of the Psb28 subunit from the Psb28-RC47 complex of T. vulcanus with its crystal structure and NMR structure.
a, b Structural comparison of the Psb28 subunit from the Psb28-RC47 complex of T. vulcanus with its crystal structure from T. elongates (PDB ID code: 3ZPN) (a) and NMR structure from Synechocystis sp. strain PCC 6803 (PDB ID code: 2KVO) (b). Psb28 from Psb28-RC47 is shown in green, and its crystal structure or NMR structure is shown in grey.
Extended Data Fig. 7 Structural comparison of the subunits between Psb28-RC47, Psb28-PSII and native PSII (PDB: 3WU2).
Subunits in Psb28-RC47, Psb28-PSII and native PSII are shown in cyan, green and grey, respectively.
Extended Data Fig. 8 Structural conflicts between D2 and Psb28, D1 and Tsl0063.
a Overall structure of D1, D2, Psb28 and Tsl0063 in Psb28-PSII and native PSII (PDB: 3WU2). b Structural conflicts between D2 and Psb28. c N-terminal region (E7-F14) of Tsl0063 (Psb28-PSII) and the D-E region (E226-G236) of D1 from native PSII. Enlarged view of the boxed area shows the amino acid residues involved in the structural conflicts. D1 subunits in Psb28-PSII and native PSII are shown in yellow and grey, respectively; D2 subunits in Psb28-PSII and native PSII are shown in marine and grey, respectively; Psb28 and Tsl0063 subunits in Psb28-PSII is shown in green and cyan, respectively.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Tables 1–3.
Source data
Source Data Extended Data Fig. 1
Unprocessed gels and western blots for Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Unprocessed gels for Extended Data Fig. 2.
Rights and permissions
About this article
Cite this article
Xiao, Y., Huang, G., You, X. et al. Structural insights into cyanobacterial photosystem II intermediates associated with Psb28 and Tsl0063. Nat. Plants 7, 1132–1142 (2021). https://doi.org/10.1038/s41477-021-00961-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00961-7
This article is cited by
-
Computational dissection of genetic variation modulating the response of multiple photosynthetic phenotypes to the light environment
BMC Genomics (2024)
-
Thermophilic cyanobacteria—exciting, yet challenging biotechnological chassis
Applied Microbiology and Biotechnology (2024)
-
The photosystem-II repair cycle: updates and open questions
Planta (2024)
-
Indirect interactions involving the PsbM or PsbT subunits and the PsbO, PsbU and PsbV proteins stabilize assembly and activity of Photosystem II in Synechocystis sp. PCC 6803
Photosynthesis Research (2024)
-
The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis
Nature Communications (2023)