Abstract
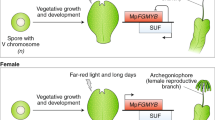
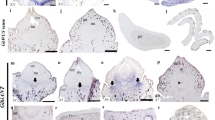
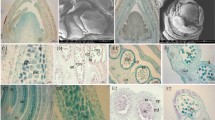
The appearance of plant organs mediated the explosive radiation of land plants, which shaped the biosphere and allowed the establishment of terrestrial animal life. The evolution of organs and immobile gametes required the coordinated acquisition of novel gene functions, the co-option of existing genes and the development of novel regulatory programmes. However, no large-scale analyses of genomic and transcriptomic data have been performed for land plants. To remedy this, we generated gene expression atlases for various organs and gametes of ten plant species comprising bryophytes, vascular plants, gymnosperms and flowering plants. A comparative analysis of the atlases identified hundreds of organ- and gamete-specific orthogroups and revealed that most of the specific transcriptomes are significantly conserved. Interestingly, our results suggest that co-option of existing genes is the main mechanism for evolving new organs. In contrast to female gametes, male gametes showed a high number and conservation of specific genes, which indicates that male reproduction is highly specialized. The expression atlas capturing pollen development revealed numerous transcription factors and kinases essential for pollen biogenesis and function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The fastq files are available for Arabidopsis (E-MTAB-9456), Amborella (E-MTAB-9190), Marchantia (E-MTAB-9457), Physcomitrium (E-MTAB-9466), maize (E-MTAB-9692) and tomato (E-MTAB-9725). The data can be obtained from https://www.ebi.ac.uk/ena.
References
Jill Harrison, C. Development and genetics in the evolution of land plant body plans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20150490 (2017).
Fürst-Jansen, J. M. R., de Vries, S. & de Vries, J. Evo-physio: on stress responses and the earliest land plants. J. Exp. Bot. 71, 3254–3269 (2020).
Brown, R. C. & Lemmon, B. E. Spores before sporophytes: hypothesizing the origin of sporogenesis at the algal–plant transition. New Phytol. 190, 875–881 (2011).
Edwards, D., Morris, J. L., Richardson, J. B. & Kenrick, P. Cryptospores and cryptophytes reveal hidden diversity in early land floras. New Phytol. 202, 50–78 (2014).
Kenrick, P. & Crane, P. R. The origin and early evolution of plants on land. Nature 389, 33–39 (1997).
Berner, R. A. GEOCARBSULF: a combined model for Phanerozoic atmospheric O2 and CO2. Geochim. Cosmochim. Acta 70, 5653–5664 (2006).
Beerling, D. J., Osborne, C. P. & Chaloner, W. G. Evolution of leaf-form in land plants linked to atmospheric CO2 decline in the Late Palaeozoic era. Nature 410, 352–354 (2001).
Menand, B. et al. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480 (2007).
Hater, F., Nakel, T. & Groß-Hardt, R. Reproductive multitasking: the female gametophyte. Annu. Rev. Plant Biol. 71, 517–546 (2020).
Hackenberg, D. & Twell, D. The evolution and patterning of male gametophyte development. Curr. Top. Dev. Biol. 131, 257–298 (2019).
Amici, G. B. Observations microscopiques sur diverses espèces de plantes. Ann. Sei. Nat. Bot. 2, 211–248 (1824).
Johnson, M. A., Harper, J. F. & Palanivelu, R. A fruitful journey: pollen tube navigation from germination to fertilization. Annu. Rev. Plant Biol. 70, 809–837 (2019).
Sprunck, S. Twice the fun, double the trouble: gamete interactions in flowering plants. Curr. Opin. Plant Biol. 53, 106–116 (2020).
Borg, M. et al. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 23, 534–549 (2011).
Favery, B. et al. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15, 79–89 (2001).
Denninger, P. et al. Male–female communication triggers calcium signatures during fertilization in Arabidopsis. Nat. Commun. 5, 4645 (2014).
Borges, F. et al. FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods 8, 44 (2012).
Borg, M. et al. An EAR-dependent regulatory module promotes male germ cell division and sperm fertility in Arabidopsis. Plant Cell 26, 2098–2113 (2014).
Cyprys, P., Lindemeier, M. & Sprunck, S. Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat. Plants 5, 253–257 (2019).
Bowles, A. M. C., Bechtold, U. & Paps, J. The origin of land plants is rooted in two bursts of genomic novelty. Curr. Biol. 30, 530–536.e2 (2020).
Rhee, S. Y. & Mutwil, M. Towards revealing the functions of all genes in plants. Trends Plant Sci. 19, 212–221 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Pina, C., Pinto, F., Feijó, J. A. & Becker, J. D. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138, 744–756 (2005).
Steffen, J. G., Kang, I.-H., Macfarlane, J. & Drews, G. N. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292 (2007).
Bowman, J. L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3, 17–22 (2000).
Kim, J. H. & Lee, B. H. GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49, 463–468 (2006).
Ding, Z. J. et al. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69 (2015).
Long, T. A. et al. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236 (2010).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Domazet-Loso, T., Brajković, J. & Tautz, D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23, 533–539 (2007).
Begun, D. J., Lindfors, H. A., Kern, A. D. & Jones, C. D. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176, 1131–1137 (2007).
Gossmann, T. I., Saleh, D., Schmid, M. W., Spence, M. A. & Schmid, K. J. Transcriptomes of plant gametophytes have a higher proportion of rapidly evolving and young genes than sporophytes. Mol. Biol. Evol. 33, 1669–1678 (2016).
Cui, X. et al. Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol. Plant 8, 935–945 (2015).
Moyers, B. A. & Zhang, J. Further simulations and analyses demonstrate open problems of phylostratigraphy. Genome Biol. Evol. 9, 1519–1527 (2017).
Doyle, J. A. in Annual Plant Reviews (eds Roberts, J. A. et al.) 1–50 (John Wiley & Sons, 2018).
Pires, N. D. & Dolan, L. Morphological evolution in land plants: new designs with old genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 508–518 (2012).
Cardona, T. Thinking twice about the evolution of photosynthesis. Open Biol. 9, 180246 (2019).
Harrison, C. J. & Morris, J. L. The origin and early evolution of vascular plant shoots and leaves. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160496 (2018).
Hetherington, A. J. & Dolan, L. Stepwise and independent origins of roots among land plants. Nature 561, 235–238 (2018).
Specht, C. D. & Bartlett, M. E. Flower evolution: the origin and subsequent diversification of the angiosperm flower. Annu. Rev. Ecol. Evol. Syst. 40, 217–243 (2009).
Pires, N. D. et al. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl Acad. Sci. USA 110, 9571–9576 (2013).
Huang, L. & Schiefelbein, J. Conserved gene expression programs in developing roots from diverse plants. Plant Cell 27, 2119–2132 (2015).
Tanabe, Y. et al. Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc. Natl Acad. Sci. USA 102, 2436–2441 (2005).
Brodribb, T. J., Carriquí, M., Delzon, S., McAdam, S. A. M. & Holbrook, N. M. Advanced vascular function discovered in a widespread moss. Nat. Plants 6, 273–279 (2020).
Ruprecht, C. et al. Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 90, 447–465 (2017).
Guo, Y.-L. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 73, 941–951 (2013).
Buschiazzo, E., Ritland, C., Bohlmann, J. & Ritland, K. Slow but not low: genomic comparisons reveal slower evolutionary rate and higher dN/dS in conifers compared to angiosperms. BMC Evol. Biol. 12, 8 (2012).
Moyle, L. C., Wu, M. & Gibson, M. J. S. Reproductive proteins evolve faster than non-reproductive proteins among Solanum species. Front. Plant Sci. 12, 635990 (2021).
Chibalina, M. V. & Filatov, D. A. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr. Biol. 21, 1475–1479 (2011).
Borg, M. et al. Epigenetic reprogramming rewires transcription during the alternation of generations in Arabidopsis. eLife 10, e61894 (2021).
Rao, X. & Dixon, R. A. Co-expression networks for plant biology: why and how. Acta Biochim. Biophys. Sin. (Shanghai) 51, 981–988 (2019).
Borges, F. et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148, 1168–1181 (2008).
Becker, J. D., Takeda, S., Borges, F., Dolan, L. & Feijó, J. A. Transcriptional profiling of Arabidopsis root hairs and pollen defines an apical cell growth signature. BMC Plant Biol. 14, 197 (2014).
von Besser, K., Frank, A. C., Johnson, M. A. & Preuss, D. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133, 4761–4769 (2006).
Proost, S. & Mutwil, M. CoNekT: an open-source framework for comparative genomic and transcriptomic network analyses. Nucleic Acids Res. 46, W133–W140 (2018).
Boisson-Dernier, A. et al. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288 (2009).
Zhu, L. et al. The Arabidopsis CrRLK1L protein kinases BUPS1 and BUPS2 are required for normal growth of pollen tubes in the pistil. Plant J. 95, 474–486 (2018).
Alves-Ferreira, M. et al. Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145, 747–762 (2007).
Gupta, R., Ting, J. T. L., Sokolov, L. N., Johnson, S. A. & Luan, S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14, 2495–2507 (2002).
Zhou, Z. et al. Arabidopsis RIC1 severs actin filaments at the apex to regulate pollen tube growth. Plant Cell 27, 1140–1161 (2015).
Liang, Y. et al. MYB97, MYB101 and MYB120 function as male factors that control pollen tube–synergid interaction in Arabidopsis thaliana fertilization. PLoS Genet. 9, e1003933 (2013).
Szövényi, P., Waller, M. & Kirbis, A. Evolution of the plant body plan. Curr. Top. Dev. Biol. 131, 1–34 (2019).
Domazet-Lošo, T. & Tautz, D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468, 815–818 (2010).
Guijarro-Clarke, C., Holland, P. W. H. & Paps, J. Widespread patterns of gene loss in the evolution of the animal kingdom. Nat. Ecol. Evol. 4, 519–523 (2020).
Xiao, S.-J., Zhang, C., Zou, Q. & Ji, Z.-L. TiSGeD: a database for tissue-specific genes. Bioinformatics 26, 1273–1275 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57, 289–300 (1995).
One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019).
Huerta-Cepas, J., Serra, F. & Bork, P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016).
Zheng, Y. et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670 (2016).
Tian, F., Yang, D.-C., Meng, Y.-Q., Jin, J. & Gao, G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2020).
Ballester, A.-R. et al. Genome, transcriptome, and functional analyses of penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol. Plant Microbe Interact. 28, 232–248 (2015).
Huerta-Cepas, J. et al. PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 39, D556–D560 (2011).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
I.J. is supported by Singaporean Ministry of Education grant MOE2018-T2-2-053, while M.M. is supported by a NTU Start-Up Grant. ERA-CAPS EVO-REPRO I2163 and FWF grant P30802 were awarded to F.B.; FCT ERA-CAPS-0001-2014 and PTDC-BIA-FBT-28484-2017 to J.D.B.; and ERA-CAPS EVO-REPRO DR 334/12-1 to S.S. and T.D. D. Hackenberg was supported by ERA-CAPS UK Biotechnology and Biological Research Council grant BB/N005090 awarded to D.T.; M.B. was supported through the FWF Lise Meitner fellowship M1818. The Vienna BioCenter Core Facilities GmbH (VBCF) Plant Sciences Facility acknowledges funding from the Austrian Federal Ministry of Education, Science and Research and the City of Vienna. L.S. was supported by CSF grant 17-23183S. C.M. and D. Honys were supported by the Czech Ministry of Education, Youth and Sport (LTC18034 and LTAIN19030) through the European Regional Development Fund-Project “Centre for Experimental Plant Biology” number CZ.02.1.01/0.0/0.0/16_019/0000738. The Genomics Unit of Instituto Gulbenkian de Ciência was partially supported by the ONEIDA Project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI–‘Fundos Europeus Estruturais e de Investimento’ from the ‘Programa Operacional Regional Lisboa 2020’ and by national funds from FCT–‘Fundação para a Ciência e a Tecnologia’. C.S.M. acknowledges a doctoral fellowship from the FCT (PD/BD/114362/2016) under the Plants for Life PhD Program. J.D.B. received salary support from the FCT through an ‘Investigador FCT’ position. M.J. and J.G. were supported by a US National Science Foundation grant (IOS-1540019). Help with sample generation was provided by L. Z. Drábková and D. Reňák. Marchantia growth was performed by the Plant Sciences Facility at the Vienna BioCenter Core Facilities GmbH (VBCF), member of the Vienna BioCenter (VBC), Austria. M. Weigend, C. Löhne and B. Reinken (Botanical Garden of the University of Bonn, Germany) are acknowledged for providing A. trichopoda plant material. D. Shivhare is acknowledged for a preliminary analysis of Physcomitrium RNA-seq data. We thank D. Maizels (http://www.scientific-art.com/) for the illustrations in Figs. 1 and 5.
Author information
Authors and Affiliations
Contributions
J.D.B. and M.M. conceived and designed the analysis. A.-C.L., M.F.-T., S.G.P., C.S.M., J.G., M.J., I.J., L.S., C.M., D. Honys and D. Hackenberg collected the data. F.B., M.B., S.S., T.D., T.K. and D.T. contributed data or analysis tools. I.J., C.F., S.P., A.-C.L. and M.M. performed the analyses. I.J., J.D.B. and M.M. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Jan de Vries, Dabing Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11 and Supplementary Methods.
Supplementary Data
Supplementary Tables 1–22.
Rights and permissions
About this article
Cite this article
Julca, I., Ferrari, C., Flores-Tornero, M. et al. Comparative transcriptomic analysis reveals conserved programmes underpinning organogenesis and reproduction in land plants. Nat. Plants 7, 1143–1159 (2021). https://doi.org/10.1038/s41477-021-00958-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00958-2
This article is cited by
-
Decoding the leaf apical meristem of Guarea glabra Vahl (Meliaceae): insight into the evolution of indeterminate pinnate leaves
Scientific Reports (2024)
-
Comparative Transcriptome Analysis Reveals the Complex Molecular Mechanisms Underlying Ultraviolet-B Tolerance in Brassica rapa var. rapa
Journal of Plant Growth Regulation (2024)
-
Cross-stress gene expression atlas of Marchantia polymorpha reveals the hierarchy and regulatory principles of abiotic stress responses
Nature Communications (2023)
-
Comparative phylotranscriptomics reveals ancestral and derived root nodule symbiosis programmes
Nature Plants (2023)
-
Conserved and non-conserved RNA–target modules in plants: lessons for a better understanding of Marchantia development
Plant Molecular Biology (2023)