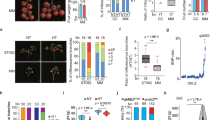
Abstract
Temperature stresses affect plant phenotypic diversity. The developmental stability of the inflorescence, required for reproductive success, is tightly regulated by the interplay of genetic and environmental factors. However, the mechanisms underpinning how plant inflorescence architecture responds to temperature are largely unknown. We demonstrate that the barley SEPALLATA MADS-box protein HvMADS1 is responsible for maintaining an unbranched spike architecture at high temperatures, while the loss-of-function mutant forms a branched inflorescence-like structure. HvMADS1 exhibits increased binding to target promoters via A-tract CArG-box motifs, which change conformation with temperature. Target genes for high-temperature-dependent HvMADS1 activation are predominantly associated with inflorescence differentiation and phytohormone signalling. HvMADS1 directly regulates the cytokinin-degrading enzyme HvCKX3 to integrate temperature response and cytokinin homeostasis, which is required to repress meristem cell cycle/division. Our findings reveal a mechanism by which genetic factors direct plant thermomorphogenesis, extending the recognized role of plant MADS-box proteins in floral development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw data files for the RNA-seq analysis reported in this paper have been deposited in the GEO database (accession no. GSE156526). The data supporting the findings of this study are available within the paper and its Supplementary Information files. Source data are provided with this paper. Additional data, such as raw image files, that support the findings of this study are available from the corresponding author upon request.
References
Franks, S. J., Sim, S. & Weis, A. E. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl Acad. Sci. USA 104, 1278–1282 (2007).
Nicotra, A. B. et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692 (2010).
Scheepens, J. F., Deng, Y. & Bossdorf, O. Phenotypic plasticity in response to temperature fluctuations is genetically variable, and relates to climatic variability of origin, in Arabidopsis thaliana. AoB Plants 10, ply043 (2018).
Casal, J. J. & Balasubramanian, S. Thermomorphogenesis. Annu. Rev. Plant Biol. 70, 321–346 (2019).
Quint, M. et al. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2, 15190 (2016).
Kumar, S. V. & Wigge, P. A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147 (2010).
Jung, J. H. et al. Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016).
Kumar, S. V. et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245 (2012).
Bommert, P. & Whipple, C. Grass inflorescence architecture and meristem determinacy. Semin. Cell Dev. Biol. 79, 37–47 (2018).
Zhang, D. & Yuan, Z. Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65, 553–578 (2014).
Wang, C., Yang, X. & Li, G. Molecular insights into inflorescence meristem specification for yield potential in cereal crops. Int. J. Mol. Sci. 22, 3508 (2021).
Prusinkiewicz, P., Erasmus, Y., Lane, B., Harder, L. D. & Coen, E. Evolution and development of inflorescence architectures. Science 316, 1452–1456 (2007).
Jacott, C. N. & Boden, S. A. Feeling the heat: developmental and molecular responses of wheat and barley to high ambient temperatures. J. Exp. Bot. 71, 5740–5751 (2020).
Urban, M. C. Accelerating extinction risk from climate change. Science 348, 571–573 (2015).
Preston, J. C. & Fjellheim, S. Understanding past, and predicting future, niche transitions based on grass flowering time variation. Plant Physiol. 183, 822–839 (2020).
Wahid, A., Gelani, S., Ashraf, M. & Foolad, M. R. Heat tolerance in plants: an overview. Environ. Exp. Bot. 61, 199–223 (2007).
Lobell, D. B., Schlenker, W. & Costa-Roberts, J. Climate trends and global crop production since 1980. Science 333, 1186–1189 (2011).
Zhao, C. et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl Acad. Sci. USA 114, 9326–9331 (2017).
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E. & Yanofsky, M. F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203 (2000).
Malcomber, S. T. & Kellogg, E. A. SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 10, 427–435 (2005).
Liu, C. et al. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell 24, 612–622 (2013).
Gao, X. C. et al. The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740 (2010).
Soyk, S. et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169, 1142–1155 (2017).
Käppel, S., Melzer, R., Rümpler, F., Gafert, C. & Theißen, G. The floral homeotic protein SEPALLATA 3 recognizes target DNA sequences by shape readout involving a conserved arginine residue in the MADS‐domain. Plant J. 95, 341–357 (2018).
Muiño, J. M., Smaczniak, C., Angenent, G. C., Kaufmann, K. & van Dijk, A. D. Structural determinants of DNA recognition by plant MADS-domain transcription factors. Nucleic Acids Res. 42, 2138–2146 (2014).
Rohs, R. et al. The role of DNA shape in protein–DNA recognition. Nature 461, 1248–1253 (2009).
Prosseda, G. et al. A temperature-induced narrow DNA curvature range sustains the maximum activity of a bacterial promoter in vitro. Biochemistry 49, 2778–2785 (2010).
Huang, Q., Duan, B., Dong, X., Fan, S. & Xia, B. GapR binds DNA through dynamic opening of its tetrameric interface. Nucleic Acids Res. 48, 9372–9386 (2020).
Callens, C., Tucker, M. R., Zhang, D. & Wilson, Z. A. Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 69, 2435–2459 (2018).
Liu, H. et al. Transcriptome profiling reveals phase-specific gene expression in the developing barley inflorescence. Crop J. 8, 71–86 (2020).
Ma, X. et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284 (2015).
Poursarebani, N. et al. COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat. Commun. 11, 5138 (2020).
Poursarebani, N. et al. The genetic basis of composite spike form in barley and ‘Miracle-Wheat’. Genetics 201, 155–165 (2015).
Koppolu, R. et al. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl Acad. Sci. USA 110, 13198–13203 (2013).
Waddington, S. R., Cartwright, P. M. & Wall, P. C. A quantitative scale of spike initial and pistil development in barley and wheat. Ann. Bot. 51, 119–130 (1983).
Ramsay, L. et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43, 169–172 (2011).
Shang, Y. et al. A CYC/TB1-type TCP transcription factor controls spikelet meristem identity in barley. J. Exp. Bot. 71, 7118–7131 (2020).
Komatsuda, T. et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl Acad. Sci. USA 104, 1424–1429 (2007).
Youssef, H. M. et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 49, 157–161 (2017).
Bull, H. et al. Barley SIX-ROWED SPIKE3 encodes a putative Jumonji C-type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nat. Commun. 8, 936 (2017).
Chuck, G., Muszynski, M., Kellogg, E., Hake, S. & Schmidt, R. J. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298, 1238–1241 (2002).
Komatsu, M., Chujo, A., Nagato, Y., Shimamoto, K. & Kyozuka, J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850 (2003).
Satoh-Nagasawa, N., Nagasawa, N., Malcomber, S., Sakai, H. & Jackson, D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441, 227–230 (2006).
Menges, M., De Jager, S. M., Gruissem, W. & Murray, J. A. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41, 546–566 (2005).
Li, X. M. et al. Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833 (2015).
Shen, H. et al. Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33, 996–1003 (2015).
Werner, T. & Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 12, 527–538 (2009).
Ashikari, M. et al. Cytokinin oxidase regulates rice grain production. Science 309, 741–745 (2005).
Han, Y., Zhang, C., Yang, H. & Jiao, Y. Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 6840–6845 (2014).
Russell, J. et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 48, 1024–1030 (2016).
Jeon, J. S. et al. leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884 (2000).
Wu, D. et al. Loss of LOFSEP transcription factor function converts spikelet to leaf-like structures in rice. Plant Physiol. 176, 1646–1664 (2018).
Cui, R. et al. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61, 767–781 (2010).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433 (2017).
Harwood, W. A. et al. in Transgenic Wheat, Barley and Oats (eds Jones, H. & Shewry, P.) 137–147 (Humana, 2009).
Zürcher, E. et al. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161, 1066–1075 (2013).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Lim, W. L. et al. Overexpression of HvCslF6 in barley grain alters carbohydrate partitioning plus transfer tissue and endosperm development. J. Exp. Bot. 71, 138–153 (2020).
Heisler, M. G. & Ohno, C. in Flower Development (eds Riechmann, J. & Wellmer, F.) 431–440 (Humana, 2014).
Kotogány, E., Dudits, D., Horváth, G. V. & Ayaydin, F. A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6, 5 (2010).
Xiong, Y. et al. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 (2013).
Li, G. et al. Rice actin-binding protein RMD is a key link in the auxin–actin regulatory loop that controls cell growth. Proc. Natl Acad. Sci. USA 111, 10377–10382 (2014).
Yang, X. et al. A rice glutamyl-tRNA synthetase modulates early anther cell division and patterning. Plant Physiol. 177, 728–744 (2018).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. (Babraham Bioinformatics, 2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
R Development Core Team. R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2017); http://www.R-project.org/
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Rau, A. & Maugis-Rabusseau, C. Transformation and model choice for RNA-seq co-expression analysis. Brief. Bioinform. 19, 425–436 (2017).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Desvoyes, B., Vergara, Z., Sequeira-Mendes, J., Madeira, S. & Gutierrez, C. in Plant Chromatin Dynamics (eds Bemer, M. & Baroux, C.) 71–82 (Humana, 2018).
Bowler, C. et al. Chromatin techniques for plant cells. Plant J. 39, 776–789 (2004).
Smaczniak, C. et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl Acad. Sci. USA 109, 1560–1565 (2012).
Du, Y. et al. UNBRANCHED3 regulates branching by modulating cytokinin biosynthesis and signaling in maize and rice. New Phytol. 214, 721–733 (2017).
Kojima, M. et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50, 1201–1214 (2009).
Acknowledgements
We thank L. Dreni (Instituto de Biología Molecular y Celular de Plantas, Spain), M. Kater (University of Milan, Italy) and M. Bennett (University of Nottingham, UK) for the discussion on the Australian Research Council Discovery Project (grant no. DP170103352 to D.Z. and R.A.B.) and Y. Liu (South China Agricultural University) for providing vectors pYLsgRNA–OsU6a, pYLsgRNA–OsU6b and pYLCRISPR–Cas9Pubi–H for CRISPR–Cas9 editing. We thank J. Chu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for help with the CK measurements, G. Mayo (Adelaide Microscopy) for assistance in the microscopy work, D. Peet (University of Adelaide) for guidance with dual-luciferase measurement, A. Chieng and H. Zhou (University of Adelaide) for managing the plant materials, and G. Coupland (Max Planck Institute for Plant Breeding Research, Germany), E. A. Kellogg (Donald Danforth Plant Science Center, USA), D. Smyth (Monash University, Australia), C. Beveridge (University of Queensland, Australia), M. Byrne (University of Sydney, Australia) and S. Boden (University of Adelaide, Australia) for comments on the manuscript. This work was financially supported by the Australian Research Council (grant no. DP170103352 to D.Z. and R.A.B.); an Australia–China Science and Research Fund Joint Research Centre grant (no. ACSRF48187) to D.Z. and G.L.; the Waite Research Institute (WRI) of the University of Adelaide; the National Key Technologies Research and Development Program of China, Ministry of Science and Technology (grant nos. 2016YFD0100804 and 2016YFE0101000 to D.Z.); the National Natural Science Foundation of China (grant nos. 31970803 and 31861163002 to D.Z.) and the Science and Research grant of Southwest University of Science and Technology (no. 19zx7146 to G.L.).
Author information
Authors and Affiliations
Contributions
D.Z. and R.A.B. initiated the project. G.L. and D.Z. conceived the project and designed the experiments. G.L. carried out most of the molecular and regulatory experiments. G.L. and H.N.J.K. created the barley sepallata mutant lines. H.N.J.K. and X.Y. conducted the scanning electron microscopy work, EdU labelling and RNA in situ hybridization. X.Y. carried out the pro::HvMADS1–eGFP transformation. H.L. and J.S. analysed the RNA-seq data under the guidance of G.L. and W.L. X.Y., C.S. and M.R.T. conducted the CK biosensor line and BAP treatment. G.L., H.N.J.K., X.Y., M.R.T., R.W., R.A.B. and D.Z. analysed the results and discussion. G.L., N.B. and D.Z. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Maria von Korff, Clinton Whipple and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Creation of barley sep mutants using CRISPR–Cas9.
a, The gene structure of HvMADS1 and positions of two sgRNA targets (T1 and T2) for CRISPR–Cas9 editing in the MADS-box domain. Blue rectangles indicate exons of HvMADS1. b, DNA sequences of independent T0 transgenics of Hvmads1 (hvm1) mutants in GP, WI, and Vla backgrounds, and hvm1/5 and hvm1/34 double mutants in GP, carrying putative HvMADS1 biallelic and homozygous mutations. WT, wild-type. c, The putative amino acid sequences encoding HvMADS1 of hvm1 single mutant, and hvm1/5 and hvm1/34 double mutants [from (b)]. Asterisks indicate a stop codon. d,e, Genotypes of three independent lines of two sgRNA targets of HvMADS5 (d) and HvMADS34 (e) in hvm5 and hvm34 single mutants, and hvm1/5, hvm1/34 and hvm5/34 double mutants that were used for CRISPR–Cas9 editing, respectively.
Extended Data Fig. 2 Spike phenotypes of sep single and double mutants under control and heat stress conditions.
Images represent spike architecture of barley WT (GP), hvm1, hvm5, hvm34 single mutants, and hvm1/5, hvm1/34, hvm5/34 double mutants at 15 °C and 28 °C. Red arrows indicate the ectopic organs. Bars = 2 cm, bars in enlarged regions are 1 cm.
Extended Data Fig. 3 Spikelet phenotype of Hvmads1 mutant under normal temperature.
a, The awn phenotype of hvm1 central spikelet in GP, Vla and WI backgrounds at 15 °C. Yellow asterisks indicate awn length. Bars = 1 cm. b, Average awn length in hvm1 and WT plants. Data shown as mean ± s.d. P values indicate results from indicated pairwise comparisons of one-way ANOVA tests. c, Floret organ (lemma, palea, stamen and pistil) phenotype in the WT (GP) and hvm1 plants at 15 °C. CS, central spikelet; LS, lateral spikelet; le, lemma; pa, palea; gl, glume; st, stamen; ca, carpel; lo, lodicule. Bars = 1 mm.
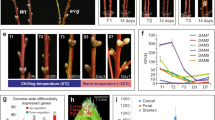
Extended Data Fig. 4 High ambient temperature induces the production of ectopic organs in Hvmads1 inflorescences.
a, WT (GP) inflorescence architecture at W9 at 28 °C. Bar = 0.5 cm. b, The developing hvm1 inflorescence from W5–9 at 28 °C. Red arrows indicate ectopic organs. Bars = 0.5 cm. c, The hvm1 heading spike at 28 °C. Red arrows indicate ectopic organs. Bar = 0.5 cm. d–f, The ES (ectopic spikelet) (d) and EI (ectopic inflorescence) (e,f) of the hvm1 spike grown at 28 °C. CS, central spikelet. Bars = 0.2 cm. g, The frequency of ES and EI in hvm1 spike sections (basal, central and apical) at different temperatures. Data shown as mean ± s.d., h, The average ES and EI number per hvm1 spike at five temperature conditions. Data shown as mean ± s.d. i, Total spikelet numbers, including spikelet from ES/EI, per WT or hvm1 spike at W7 at different temperatures. Data shown as mean ± s.d. P values indicate results from indicated pairwise comparisons of one-way ANOVA tests. j,k, The ES and EI induced by high ambient temperatures in hvm1 mutants of Vla (j) and WI (k) backgrounds. Bars = 0.5 cm. l,m, short (l) and elongated (m) EI branches with different spikelet morphology. s-EI, secondary EI branch; LS, lateral spikelet; le, lemma; pa, palea; st, stamen; ca, carpel; gl, glume. Bars = 0.5 cm. n, The frequency of short and elongated EI phenotype in hvm1 mutants at different temperatures. o, Mature spike of hvm1 (GP) mutant after treatment at high temperatures. Red arrows indicate fertile spikelets from EI or ES. Bar = 1 cm. p, Spikelet fertility rate of EI induced by high temperatures in hvm1 (GP) spikes. Data shown as mean ± s.d. q, Fertility rate of ES in hvm1 mutants of three backgrounds at 23 °C and 28 °C. Data shown as mean ± s.d. All individual biological experiments were repeated at least three times with similar results.
Extended Data Fig. 5 Loss of Hvmads1 leads to reduced meristem determinacy and delayed inflorescence development under high temperature.
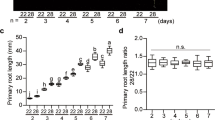
a,b, Scanning electron microscopy of spike morphology at W2.5, W3.5 and W7 in WT (GP) (a) and hvm1 (b) plants at 15 °C, showing the short awn in hvm1. c, Morphology of the developing WT spike at 28 °C. d, Reduced meristem determinacy of hvm1 inflorescences at 28 °C. Green asterisks indicate the indeterminate inflorescence meristem likely converted from the central spikelet meristems, yellow asterisks indicate the ectopic initiated meristems or inflorescence/spikelet meristems possibly reverted from rachilla. e, Effects of ambient high temperatures 20 °C, 23 °C and 25 °C on morphology of hvm1 spike. Yellow shading indicates EI, blue shading indicates ES. All bars (a–e) = 100 µm. fm, floral meristem; ls, lateral spikelet; cs, central spikelet; gl, glume; st, stamen; pi, pistil; le, lemma; esm, ectopic spikelet meristem; eim, ectopic inflorescence meristem. f, Rate of spike development at different temperatures, showing delay in hvm1 (GP) at 28 °C, compared with WT, spike development. Bars = 0.5 mm. g, Days to reach different Waddington stages of spike development at 15 °C and 28 °C in three barley varieties and related hvm1 mutants. Data shown as mean ± s.d. P < 0.001, two-way ANOVA tests of WT and hvm1 (three background comparisons) at 28 °C. All experiments with treatment were repeated independently at least three times with similar results.
Extended Data Fig. 6 HvMADS1 represses cell division in the spike in response to high temperature.
a, Indicative method of EdU (5-ethynyl-2’-deoxyuridine) tracking in barley spike. PI, propidium iodide. b,c, EdU tracking of cell division activities in WT (GP) and hvm1 spikes (W2.5 and W3–3.5) grown at 15 °C (b) and 25 °C (c). White arrows indicate ectopic EdU clusters in non-floret meristem regions of the central spikelet, which shows high levels of cell division activity in ectopic meristems of hvm1 plants. Numbers of ectopic clusters represent the average observed additional EdU signal clusters in non-floret meristem regions per spike (W3–3.5). Also see the Source Data. Bars = 100 µm. The pictures of EdU tracking assays represent one of three experiments performed independently with similar results.
Extended Data Fig. 7 Temperature does not alter HvMADS1 mRNA expression or protein accumulation.
a, Accumulation of the HvMADS1 protein in flower organs, including lemma, palea, anther, and lodicule, in pro::HvMADS1-eGFP transgenic lines at 15 °C. BF, bright field. Bars = 100 µm. b, qRT-PCR analysis of HvMADS1 expression in W2.5 and W3.5 WT spikes at different temperatures. A temperature-responsive gene (HB, homeobox) served as the positive control. Data shown as mean ± s.d., n = 3 biological replicates. c, RT-PCR analysis of HvMADS1 expression in WT spikes (W3.5). HvActin7 served as the control. kb indicates kilobase. d, Immunoblot analysis of HvMADS1-eGFP protein in W3.5 spikes from three independent pro::HvMADS1-eGFP transgenic lines in response to temperatures. Tubulin served as a loading control. All experiments were repeated independently at least three times with similar results.
Extended Data Fig. 8 HvMADS1 regulates the transcriptome of barley inflorescence in response to temperature.
a, Correlation analysis of transcripts showing mis-regulation of spike developmental genes in hvm1 plants at 15 °C (left) and, more obviously, at 25 °C (right). b, Venn diagram showing the number of DEGs affected by genotype and temperature at two developmental stages. c, Co-expression clustering of all DEGs in eight transcriptomes (W2.5 and W3.5 of WT and hvm1 spikes at 15 °C and 25 °C). Clustering was performed on the expression-filtered data set using a Gaussian mixture model. The number of clusters was assumed to be random and was automatically learned using an empirical Bayes approach (variational Bayesian inference). d, DEG clusters in response to temperature, developmental phase and HvMADS1 genotype in inflorescence meristems. Z-score represents variation in gene expression that is likely to be regulated by the interaction between phase × temperature × genotype, temperature × genotype, or only HvMADS1 genotype for selected clusters. Three biological repeats were performed for transcriptome.
Extended Data Fig. 9 Effects of Hvmads1 mutation on the expression of key regulators in response to temperature.
a, Heat map showing DEGs relevant to spike development (left) and temperature response (right). b, qRT-PCR analysis of selected genes related to inflorescence meristem identity in W3.5 WT (GP) and hvm1 spikes at 15 °C and 25 °C. OSH1, ORYZA SATIVA HOMEOBOX1; AP1, APETALA 1; VRN, VERNALIZATION; TFL1, TERMINAL FLOWER 1; TB1, TEOSINTE BRANCHED 1; TAW1, TAWAWA1; RPK4, RECEPTOR-LIKE PROTEIN KINASE 4; IDS1, INDETERMINATE SPIKELET 1. c, qRT-PCR analysis of selected genes known to regulate barley spike development, spikelet identity and row-type in W3.5 WT and hvm1 spikes at 15 °C and 25 °C. VRS, SIX-ROWED SPIKE; COM2, COMPOSITUM 2. d, qRT-PCR analysis of selected genes related to temperature response in W3.5 WT and hvm1 spikes at 15 °C and 25 °C. PIF4, PHYTOCHROME-INTERACTING FACTOR 4; ER, ERECTA; TT1, THERMO-TOLERANCE 1. HvActin7 and HvEF2 were used for normalisation. For (b–d), data shown as mean ± s.d., n = 3 biological replicates. P values indicate results from indicated pairwise comparisons of one-way ANOVA tests (b–d).
Extended Data Fig. 10 HvMADS1 promotes the activity of the HvCKX3 promoter in a temperature-dependent manner.
Truncated HvCKX3 promoter fragments containing 0, 1, 2 or 3 CArG-boxes were fused to the LUC reporter gene, and co-transformed with effector plasmids of EV (empty vector) and 35S::HvMADS1 into tobacco cells. Normalised LUC/REN activity is shown as mean ± s.d., n = 5 biological replicates. P values indicate results from indicated pairwise comparisons of one-way ANOVA tests.
Supplementary information
Supplementary Information
Supplementary Data 1, Fig. 1, Tables 1–10 and legends for Supplementary Datasets 1–7.
Supplementary Dataset 1
DEGs across temperature, genotype and developmental phase.
Supplementary Dataset 2
Co-expression clusters of DEGs from RNA-seq of eight barley spike samples.
Supplementary Dataset 3
GO analysis of DEGs in wild-type and Hvmads1 spikes at 15 °C.
Supplementary Dataset 4
GO analysis of DEGs in wild-type and Hvmads1 spikes at 25 °C.
Supplementary Dataset 5
Curated list of genes for inflorescence development, temperature response, cell cycle/division and plant hormone pathways.
Supplementary Dataset 6
SNPs of HvMADS1 exons in 267 barley varieties.
Supplementary Dataset 7
HvMADS1 sequence variation in barley varieties.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots and gels.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Li, G., Kuijer, H.N.J., Yang, X. et al. MADS1 maintains barley spike morphology at high ambient temperatures. Nat. Plants 7, 1093–1107 (2021). https://doi.org/10.1038/s41477-021-00957-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00957-3
This article is cited by
-
MADS1-regulated lemma and awn development benefits barley yield
Nature Communications (2024)
-
The auxin-responsive transcription factor SlDOF9 regulates inflorescence and flower development in tomato
Nature Plants (2022)