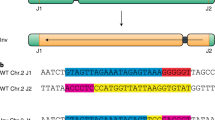
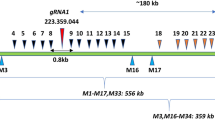
Abstract
Plant breeding relies on the presence of genetic variation, as well as on the ability to break or stabilize genetic linkages between traits. The development of the genome-editing tool clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein (Cas) has allowed breeders to induce genetic variability in a controlled and site-specific manner, and to improve traits with high efficiency. However, the presence of genetic linkages is a major obstacle to the transfer of desirable traits from wild species to their cultivated relatives. One way to address this issue is to create mutants with deficiencies in the meiotic recombination machinery, thereby enhancing global crossover frequencies between homologous parental chromosomes. Although this seemed to be a promising approach at first, thus far, no crossover frequencies could be enhanced in recombination-cold regions of the genome. Additionally, this approach can lead to unintended genomic instabilities due to DNA repair defects. Therefore, efforts have been undertaken to obtain predefined crossovers between homologues by inducing site-specific double-strand breaks (DSBs) in meiotic, as well as in somatic plant cells using CRISPR–Cas tools. However, this strategy has not been able to produce a substantial number of heritable homologous recombination-based crossovers. Most recently, heritable chromosomal rearrangements, such as inversions and translocations, have been obtained in a controlled way using CRISPR–Cas in plants. This approach unlocks a completely new way of manipulating genetic linkages, one in which the DSBs are induced in somatic cells, enabling the formation of chromosomal rearrangements in the megabase range, by DSB repair via non-homologous end-joining. This technology might also enable the restructuring of genomes more globally, resulting in not only the obtainment of synthetic plant chromosome, but also of novel plant species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D. & Schroeder, J. I. Genetic strategies for improving crop yields. Nature 575, 109–118 (2019).
Wolter, F., Schindele, P. & Puchta, H. Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19, 176 (2019).
Lewis, R. S. & Rose, C. Agronomic performance of tobacco mosaic virus-resistant tobacco lines and hybrids possessing the resistance gene N introgressed on different chromosomes. Crop Sci. 50, 1339–1347 (2010).
Li, J., Chitwood, J., Menda, N., Mueller, L. & Hutton, S. F. Linkage between the I-3 gene for resistance to Fusarium wilt race 3 and increased sensitivity to bacterial spot in tomato. Theor. Appl. Genet. 131, 145–155 (2018).
Zhang, Y., Pribil, M., Palmgren, M. & Gao, C. A CRISPR way for accelerating improvement of food crops. Nat. Food 1, 200–205 (2020).
Zaidi, S. S.-E.-A., Mahas, A., Vanderschuren, H. & Mahfouz, M. M. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 21, 289 (2020).
Schindele, A., Dorn, A. & Puchta, H. CRISPR/Cas brings plant biology and breeding into the fast lane. Curr. Opin. Biotechnol. 61, 7–14 (2020).
Atkins, P. A. & Voytas, D. F. Overcoming bottlenecks in plant gene editing. Curr. Opin. Plant Biol. 54, 79–84 (2020).
Zhang, Y., Malzahn, A. A., Sretenovic, S. & Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794 (2019).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Puchta, H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14 (2004).
Schmidt, C., Schindele, P. & Puchta, H. From gene editing to genome engineering: restructuring plant chromosomes via CRISPR/Cas. aBIOTECH 1, 21–31 (2020).
Rönspies, M., Schindele, P. & Puchta, H. CRISPR/Cas-mediated chromosome engineering: opening up a new avenue for plant breeding. J. Exp. Bot. 72, 177–183 (2021).
Thompson, S. L. & Compton, D. A. Chromosomes and cancer cells. Chromosome Res. 19, 433–444 (2011).
Park, C.-Y., Sung, J. J. & Kim, D.-W. Genome editing of structural variations: modeling and gene correction. Trends Biotechnol. 34, 548–561 (2016).
Brunet, E. & Jasin, M. Induction of chromosomal translocations with CRISPR-Cas9 and other nucleases: understanding the repair mechanisms that give rise to translocations. Adv. Exp. Med. Biol. 1044, 15–25 (2018).
Hasty, P. & Montagna, C. Chromosomal rearrangements in cancer: detection and potential causal mechanisms. Mol. Cell. Oncol. 1, e29904 (2014).
Lakich, D., Kazazian, H. H. Jr, Antonarakis, S. E. & Gitschier, J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat. Genet. 5, 236–241 (1993).
Bondeson, M. L. et al. Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum. Mol. Genet. 4, 615–621 (1995).
Small, K., Iber, J. & Warren, S. T. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat. Genet. 16, 96–99 (1997).
Park, C.-Y., Lee, D. R., Sung, J. J. & Kim, D.-W. Genome-editing technologies for gene correction of hemophilia. Hum. Genet. 135, 977–981 (2016).
Kirkpatrick, M. & Barton, N. Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434 (2006).
Fransz, P. et al. Molecular, genetic and evolutionary analysis of a paracentric inversion in Arabidopsis thaliana. Plant J. 88, 159–178 (2016).
Harewood, L. & Fraser, P. The impact of chromosomal rearrangements on regulation of gene expression. Hum. Mol. Genet. 23, R76–R82 (2014).
Martin, G. et al. Chromosome reciprocal translocations have accompanied subspecies evolution in bananas. Plant J. 104, 1698–1711 (2020).
Wellenreuther, M. & Bernatchez, L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 33, 427–440 (2018).
Beying, N., Schmidt, C., Pacher, M., Houben, A. & Puchta, H. CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 6, 638–645 (2020).
Schmidt, C. et al. Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. Nat. Commun. 11, 4418 (2020).
Schwartz, C. et al. CRISPR–Cas9-mediated 75.5-Mb inversion in maize. Nat. Plants 6, 1427–1431 (2020).
Taagen, E., Bogdanove, A. J. & Sorrells, M. E. Counting on crossovers: controlled recombination for plant breeding. Trends Plant Sci. 25, 455–465 (2020).
Wang, Y. & Copenhaver, G. P. Meiotic recombination: mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609 (2018).
Lambing, C., Franklin, F. C. H. & Wang, C.-J. R. Understanding and manipulating meiotic recombination in plants. Plant Physiol. 173, 1530–1542 (2017).
Bergerat, A. et al. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386, 414–417 (1997).
Crismani, W. et al. FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012).
Hartung, F., Suer, S. & Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 104, 18836–18841 (2007).
Séguéla-Arnaud, M. et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl Acad. Sci. USA 112, 4713–4718 (2015).
Dorn, A. et al. The topoisomerase 3α zinc-finger domain T1 of Arabidopsis thaliana is required for targeting the enzyme activity to Holliday junction-like DNA repair intermediates. PLoS Genet. 14, e1007674 (2018).
Séguéla-Arnaud, M. et al. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 45, 1860–1871 (2017).
de Maagd, R. A. CRISPR/Cas inactivation of RECQ4 increases homeologous crossovers in an interspecific tomato hybrid. Plant Biotechnol. J. 18, 805–813 (2020).
Girard, C. et al. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11, e1005369 (2015).
Fernandes, J. B. et al. FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet. 14, e1007317 (2018).
Zhang, P. et al. The rice AAA-ATPase OsFIGNL1 is essential for male meiosis. Front. Plant Sci. 8, 1639 (2017).
Mieulet, D. et al. Unleashing meiotic crossovers in crops. Nat. Plants 4, 1010–1016 (2018).
Pyatnitskaya, A., Borde, V. & de Muyt, A. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma 128, 181–198 (2019).
Ziolkowski, P. A. et al. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 31, 306–317 (2017).
Serra, H. et al. Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl Acad. Sci. USA 115, 2437–2442 (2018).
Wang, K., Wang, C., Liu, Q., Liu, W. & Fu, Y. Increasing the genetic recombination frequency by partial loss of function of the synaptonemal complex in rice. Mol. Plant 8, 1295–1298 (2015).
Fernandes, J. B., Séguéla-Arnaud, M., Larchevêque, C., Lloyd, A. H. & Mercier, R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl Acad. Ssci. USA 115, 2431–2436 (2018).
Hartung, F., Suer, S., Knoll, A., Wurz-Wildersinn, R. & Puchta, H. Topoisomerase 3α and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4, e1000285 (2008).
Mannuss, A. et al. RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22, 3318–3330 (2010).
Schröpfer, S., Kobbe, D., Hartung, F., Knoll, A. & Puchta, H. Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res. 42, 1684–1697 (2014).
Knoll, A. et al. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24, 1448–1464 (2012).
Peciña, A. et al. Targeted stimulation of meiotic recombination. Cell 111, 173–184 (2002).
Sarno, R. et al. Programming sites of meiotic crossovers using Spo11 fusion proteins. Nucleic Acids Res. 45, e164 (2017).
Yelina, N. E., Gonzalez-Jorge, S., Hirsz, D., Yang, Z. & Henderson, I. R. CRISPR targeting of MEIOTIC-TOPOISOMERASE VIB-dCas9 to a recombination hotspot is insufficient to increase crossover frequency in Arabidopsis. Preprint at bioRxiv https://doi.org/10.1101/2021.02.01.429210 (2021).
Filler Hayut, S., Melamed Bessudo, C. & Levy, A. A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 8, 15605 (2017).
Ben Shlush, I. et al. CRISPR/Cas9 induced somatic recombination at the CRTISO locus in tomato. Genes 12, 59 (2020).
Davis, L., Khoo, K. J., Zhang, Y. & Maizels, N. POLQ suppresses interhomolog recombination and loss of heterozygosity at targeted DNA breaks. Proc. Natl Acad. Sci. USA 117, 22900–22909 (2020).
Cheong, T.-C., Blasco, R. B. & Chiarle, R. The CRISPR/Cas9 System as a tool to engineer chromosomal translocation in vivo. Adv. Exp. Med. Biol. 1044, 39–48 (2018).
Torres-Ruiz, R. et al. Efficient recreation of t(11;22) EWSR1-FLI1+ in human stem cells Using CRISPR/Cas9. Stem Cell Rep. 8, 1408–1420 (2017).
Torres, R. et al. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR–Cas9 system. Nat. Commun. 5, 3964 (2014).
Lagutina, I. V. et al. Modeling of the human alveolar rhabdomyosarcoma Pax3–Foxo1 chromosome translocation in mouse myoblasts using CRISPR–Cas9 nuclease. PLoS Genet. 11, e1004951 (2015).
Choi, P. S. & Meyerson, M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 5, 3728 (2014).
Luo, J., Sun, X., Cormack, B. P. & Boeke, J. D. Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature 560, 392–396 (2018).
Shao, Y. et al. Creating a functional single-chromosome yeast. Nature 560, 331–335 (2018).
Fleiss, A. et al. Reshuffling yeast chromosomes with CRISPR/Cas9. PLoS Genet. 15, e1008332–e1008332 (2019).
Yadav, V., Sun, S., Coelho, M. A. & Heitman, J. Centromere scission drives chromosome shuffling and reproductive isolation. Proc. Natl Acad. Sci. USA 117, 7917–7928 (2020).
Jayakodi, M. et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284–289 (2020).
Walkowiak, S. et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 588, 277–283 (2020).
Crow, T. et al. Gene regulatory effects of a large chromosomal inversion in highland maize. PLoS Genet. 16, e1009213 (2020).
Schmidt, C., Pacher, M. & Puchta, H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 98, 577–589 (2019).
Fransz, P. F. et al. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100, 367–376 (2000).
Drouaud, J. et al. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 16, 106–114 (2006).
Zhang, Y., Cheng, Z., Ma, J., Xian, F. & Zhang, X. Characteristics of a novel male–female sterile watermelon (Citrullus lanatus) mutant. Sci. Horticulturae 140, 107–114 (2012).
Steinert, J., Schiml, S., Fauser, F. & Puchta, H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 84, 1295–1305 (2015).
Schindele, P. & Puchta, H. Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol. J. 18, 1118–1120, https://doi.org/10.1111/pbi.13275 (2020).
Wolter, F., Klemm, J. & Puchta, H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 94, 735–746 (2018).
Mandáková, T. & Lysak, M. A. Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol. 42, 55–65 (2018).
Liu, Y. et al. Rapid birth or death of centromeres on fragmented chromosomes in maize. Plant Cell 32, 3113–3123 (2020).
Wimmer, E., Mueller, S., Tumpey, T. M. & Taubenberger, J. K. Synthetic viruses: a new opportunity to understand and prevent viral disease. Nat. Biotechnol. 27, 1163–1172 (2009).
Coradini, A. L. V., Hull, C. B. & Ehrenreich, I. M. Building genomes to understand biology. Nat. Commun. 11, 6177 (2020).
Dawe, R. K. Charting the path to fully synthetic plant chromosomes. Exp. Cell. Res. 390, 111951 (2020).
Liu, J. et al. Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 31, 368–383 (2019).
Zhang, H. et al. Stable integration of an engineered megabase repeat array into the maize genome. Plant J. 70, 357–365 (2012).
Acknowledgements
We thank D. Donahey for proofreading the manuscript. This work was supported by the European Research Council (Advanced grant: ERC-2016-AdG_741306 CRISBREED).
Author information
Authors and Affiliations
Contributions
All authors wrote the manuscript. A.D. and P.S. designed the figures. P.S. created the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Huanbin Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rönspies, M., Dorn, A., Schindele, P. et al. CRISPR–Cas-mediated chromosome engineering for crop improvement and synthetic biology. Nat. Plants 7, 566–573 (2021). https://doi.org/10.1038/s41477-021-00910-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00910-4
This article is cited by
-
A designer synthetic chromosome fragment functions in moss
Nature Plants (2024)
-
Integrating machine learning and genome editing for crop improvement
aBIOTECH (2024)
-
Crop bioengineering via gene editing: reshaping the future of agriculture
Plant Cell Reports (2024)
-
Genome-wide expansion and reorganization during grass evolution: from 30 Mb chromosomes in rice and Brachypodium to 550 Mb in Avena
BMC Plant Biology (2023)
-
Hidden prevalence of deletion-inversion bi-alleles in CRISPR-mediated deletions of tandemly arrayed genes in plants
Nature Communications (2023)