Abstract
The nitrate transporter NRT2.1, which plays a central role in high-affinity nitrate uptake in roots, is activated at the post-translational level in response to nitrogen (N) starvation1,2. However, the critical enzymes required for the post-translational activation of NRT2.1 remain to be identified. Here, we show that a type 2C protein phosphatase, designated CEPD-induced phosphatase (CEPH), activates high-affinity nitrate uptake by directly dephosphorylating Ser501 of NRT2.1, a residue that functions as a negative phospho-switch in Arabidopsis2. CEPH is predominantly expressed in epidermal and cortex cells in roots and is upregulated by N starvation via a CEPDL2/CEPD1/2-mediated long-distance signalling from shoots3,4. The loss of CEPH leads to marked decreases in high-affinity nitrate uptake, tissue nitrate content and plant biomass. Collectively, our results identify CEPH as a crucial enzyme in the N-starvation-dependent activation of NRT2.1 and provide molecular and mechanistic insights into how plants regulate high-affinity nitrate uptake at the post-translational level in response to the N environment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The microarray data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE160649. The raw MS data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository under accession number PXD022343. The Arabidopsis lines generated in this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Laugier, E. et al. Regulation of high-affinity nitrate uptake in roots of Arabidopsis depends predominantly on posttranscriptional control of the NRT2.1/NAR2.1 transport system. Plant Physiol. 158, 1067–1078 (2012).
Jacquot, A. et al. NRT2.1 C-terminus phosphorylation prevents root high affinity nitrate uptake activity in Arabidopsis thaliana. N. Phytol. 228, 1038–1054 (2020).
Ohkubo, Y., Tanaka, M., Tabata, R., Ogawa-Ohnishi, M. & Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3, 17029 (2017).
Ota, R., Ohkubo, Y., Yamashita, Y., Ogawa-Ohnishi, M. & Matsubayashi, Y. Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 11, 641 (2020).
Wang, Y. Y., Cheng, Y. H., Chen, K. E. & Tsay, Y. F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122 (2018).
Wang, R. & Crawford, N. M. Genetic identification of a gene involved in constitutive, high-affinity nitrate transport in higher plants. Proc. Natl Acad. Sci. USA 93, 9297–9301 (1996).
Filleur, S. et al. An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489, 220–224 (2001).
Kiba, T. et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24, 245–258 (2012).
Tsay, Y. F., Schroeder, J. I., Feldmann, K. A. & Crawford, N. M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713 (1993).
Huang, N. C., Liu, K. H., Lo, H. J. & Tsay, Y. F. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392 (1999).
Liu, K. H., Huang, C. Y. & Tsay, Y. F. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874 (1999).
Malagoli, P. et al. Modeling nitrogen uptake in oilseed rape cv Capitol during a growth cycle using influx kinetics of root nitrate transport systems and field experimental data. Plant Physiol. 134, 388–400 (2004).
Cerezo, M. et al. Major alterations of the regulation of root NO3− uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127, 262–271 (2001).
Lejay, L. et al. Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J. 18, 509–519 (1999).
Gansel, X., Munos, S., Tillard, P. & Gojon, A. Differential regulation of the NO3− and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J. 26, 143–155 (2001).
Fraisier, V., Gojon, A., Tillard, P. & Daniel-Vedele, F. Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J. 23, 489–496 (2000).
Wirth, J. et al. Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J. Biol. Chem. 282, 23541–23552 (2007).
Jacquot, A., Li, Z., Gojon, A., Schulze, W. & Lejay, L. Post-translational regulation of nitrogen transporters in plants and microorganisms. J. Exp. Bot. 68, 2567–2580 (2017).
Engelsberger, W. R. & Schulze, W. X. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 69, 978–995 (2012).
Menz, J., Li, Z., Schulze, W. X. & Ludewig, U. Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J. 88, 717–734 (2016).
Zou, X., Liu, M. Y., Wu, W. H. & Wang, Y. Phosphorylation at Ser28 stabilizes the Arabidopsis nitrate transporter NRT2.1 in response to nitrate limitation. J. Integr. Plant Biol. 62, 865–876 (2020).
Ohyama, K., Ogawa, M. & Matsubayashi, Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC–MS-based structure analysis. Plant J. 55, 152–160 (2008).
Tabata, R. et al. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346 (2014).
Fuchs, S., Grill, E., Meskiene, I. & Schweighofer, A. Type 2C protein phosphatases in plants. FEBS J. 280, 681–693 (2013).
Bhaskara, G. B., Wong, M. M. & Verslues, P. E. The flip side of phospho-signalling: regulation of protein dephosphorylation and the protein phosphatase 2Cs. Plant Cell Environ. 42, 2913–2930 (2019).
Wang, Y. F. et al. Genome-wide identification and expression analysis of StPP2C gene family in response to multiple stresses in potato (Solarium tuberosum L.). J. Integr. Agric. 19, 1609–1624 (2020).
Yang, Q. et al. Genome-wide identification of PP2C genes and their expression profiling in response to drought and cold stresses in Medicago truncatula. Sci. Rep. 8, 12841 (2018).
Xue, T. T. et al. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9, 550 (2008).
Fan, K. et al. Molecular evolution and lineage-specific expansion of the PP2C family in Zea mays. Planta 250, 1521–1538 (2019).
Komatsu, K. et al. Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol. Biol. 70, 327–340 (2009).
Nelson, C. J., Huttlin, E. L., Hegeman, A. D., Harms, A. C. & Sussman, M. R. Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7, 1279–1292 (2007).
Bhaskara, G. B., Wen, T. N., Nguyen, T. T. & Verslues, P. E. Protein phosphatase 2Cs and Microtubule-Associated Stress Protein 1 control microtubule stability, plant growth, and drought response. Plant Cell 29, 169–191 (2017).
Kleinboelting, N., Huep, G., Kloetgen, A., Viehoever, P. & Weisshaar, B. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40, D1211–D1215 (2012).
Zuo, J., Niu, Q. W. & Chua, N. H. Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 (2000).
Wessel, D. & Flugge, U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984).
Acknowledgements
We thank H. Fukuda (Agilent Technologies) for technical advice regarding phosphopeptide enrichment. This research was supported by a Grant-in-Aid for Scientific Research (S) (no. 18H05274 to Y.M.), a Grant-in-Aid for Transformative Research Areas (A) (no. 20H05907 to Y.M.) and a Grant-in-Aid for JSPS Fellows (no. 20J20049 to Y.O.) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Contributions
Y.M. and Y.O. conceived the project and designed the experiments. Y.O. performed all of the biological experiments. K.K. performed the phosphopeptide enrichment and nano LC–MS/MS analyses. Y.M. and Y.O. interpreted the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Brent Kaiser and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
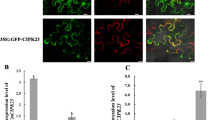
Extended Data Fig. 1 At4g32950 is an Arabidopsis PP2C subfamily E protein.
a, A combined phylogenetic tree of PP2C subfamily E proteins from Arabidopsis (At), potato (St), Medicago truncatula (Mt), rice (Os), maize (Zm) and moss Physcomitrella patens (Pp). At4g32950 was classified into a distinct clade (shaded) that includes members from potato, Medicago, rice and maize, but not mosses. Numbers represent bootstrap values from 1,000 replicates. b, Expression stability of two different reference genes, EF-1α and TUA4, between WT and cepd1,2 cepdl2 triple mutant plants. TUA4 and EF-1α expression level in WT and cepd1,2 cepdl2 was quantified by RT-qPCR using EF-1α and TUA4 as reference genes, respectively. c, Expression of NRT2.1 in WT and cepd1,2 cepdl2 triple mutant plants. Data are presented as the mean ± SD, and each dot represents a biological replicate. P-values are 2.76 × 10−2 for 3 mM and 1.61 × 10−4 for 1 mM. For (b) and (c), statistical significance is determined by two-tailed non-paired Student’s t test (*P < 0.05, ‘ns’ indicates not significant).
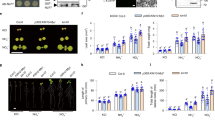
Extended Data Fig. 2 Phenotypic analysis of ceph-1 mutant plants.
a, Fresh weight of 14-day-old shoots of WT, ceph-1 mutant, and ceph-1 plants complemented with CEPH grown on 1 mM NO3– medium. b, Fresh weight of 14-day-old shoots of WT, ceph-1 mutant, and ceph-1 plants complemented with CEPH-GFP grown on 1 mM NO3– medium. c, Root phenotypes of 10-day-old WT and ceph-1 mutant plants grown on 1 mM NO3– medium. Scale bar = 1 cm. d, Primary root length of 10- or 14-day-old WT and ceph-1 mutant plants grown on 1 mM NO3– medium. e, Ratio of total lateral root (LR) length to primary root (PR) length of 10- or 14-day-old WT and ceph-1 mutant plants grown on 1 mM NO3– medium. P-values are 2.23 × 10−2 for 10 d and 4.01 × 10−3 for 14 d. f, Phenotypes of 14-day-old WT and ceph-1 mutant plants grown under N-replete conditions (10 mM NH4+, 10 mM NO3–). Scale bar = 5 mm. g, Fresh weight of the shoots and roots described in (f). (h) HATS and calculated LATS NO3– uptake activities of 14-day-old WT and ceph-1 mutant plants grown on 1 mM NO3– medium. P-values are 6.13 × 10−5 for HATS and 1.32 × 10−3 for LATS. i, Expression of nitrate transporter genes in WT and ceph-1 mutant plants. j, Expression of CEPD1/2 and CEPDL2 in WT and ceph-1 mutant plants. P-value is 3.81 × 10−3 for CEPDL2. k, Induction level of CEPH in XVE-CEPH plants 6 h after estradiol treatment. P-value is 3.65 × 10−3. Data are presented as the mean ± SD, and each dot represents a biological replicate. For (a) and (b), different letters indicate statistically significant differences (P < 0.05, one-way ANOVA with post-hoc Tukey’s test). For (d), (e) and (g)-(k), statistical significance is determined by two-tailed non-paired Student’s t test (*P < 0.05, ‘ns’ indicates not significant).
Extended Data Fig. 3 Venn diagram showing overlap between data sets.
a, Venn diagram for numbers of phosphopeptide ion peaks identified in the Lys-C–digested samples and overlap between the forward and reciprocal analyses. b, Venn diagram for numbers of phosphopeptide ion peaks identified in the trypsin-digested samples and overlap between the forward and reciprocal analyses.
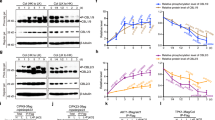
Extended Data Fig. 4 Quantitative phosphoproteomic analysis of the CEPH substrate.
a, Mass spectra of the NRT2.1 G478 nonphosphorylated peptide (position 478-493 in NRT2.1 protein) derived from reciprocally 14N- and 15N-labeled plants, showing comparable NRT2.1 protein abundance in WT and ceph-1 plants. b, Mass spectra of the NRT2.1 T521 phosphopeptide (position 516-530 in NRT2.1) derived from reciprocally labeled plants, showing comparable phosphorylation levels in WT and ceph-1 plants. (c) Dephosphorylation of the NRT2.1 S28 phosphopeptide with CEPH-GFP detected by LC-MS in the selected ion monitoring mode at m/z 831.9.
Extended Data Fig. 5 NRT2.1 S501 phosphorylation acts as a negative phospho-switch that represses HATS activity.
a, Phenotypes of 14-day-old WT, nrt2.1-2.2 mutant, nrt2.1-2.2 complemented with NRT2.1[S501A] and nrt2.1-2.2 complemented with NRT2.1[S501D] grown on 1 mM NO3– medium. Scale bar = 2 mm. b, Fresh weight of 14-day-old shoots of the plants shown in (a). c, Root HATS activity of the plants shown in (a). d, Mass spectra of the NRT2.1 G478 nonphosphorylated peptide (position 478-493 in NRT2.1 protein) and ACT8 A21 peptide (position 21-30 in ACT8 protein) derived from WT and NRT2.1[S501D] plants. Decrease in ACT8 protein abundance in S501D plants suggests the decrease in overall protein expression level is due to a nitrate uptake defect in S501D plants. Considering this, protein abundance of NRT2.1 was not significantly affected by the S501D mutation. e, Root HATS activity of 14-day-old nrt2.1-2.2 mutant plants complemented with NRT2.1[S501A], ceph-1 mutant, ceph-1 mutant expressing NRT2.1[S501A] and ceph-1 mutant expressing NRT2.1[S501D]. The plotted S501A/nrt2.1-2.2 data is the same as that shown in Extended Data Fig. 5c, as the experiments were done concurrently. f, Mass spectra of the NRT2.1 G478 nonphosphorylated peptide (position 478-493 in NRT2.1 protein) derived from reciprocally 14N- and 15N-labeled plants, showing comparable NRT2.1 protein abundance in mock and estradiol-treated XVE-CEPH plants. Data are presented as the mean ± SD, and each dot represents a biological replicate. For (b), (c) and (e), different letters indicate statistically significant differences (P < 0.05, one-way ANOVA with post-hoc Tukey’s test).
Supplementary information
Supplementary Table 1
List of the top ten CEPDL2/CEPD1/2-regulated genes in roots. The table includes the GeneChip signal intensity, signal intensity ratio (log2(FC)), z score and gene description. The results are sorted by the z score of CEPDL2ox plants.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Ohkubo, Y., Kuwata, K. & Matsubayashi, Y. A type 2C protein phosphatase activates high-affinity nitrate uptake by dephosphorylating NRT2.1. Nat. Plants 7, 310–316 (2021). https://doi.org/10.1038/s41477-021-00870-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00870-9
This article is cited by
-
IMA peptides regulate root nodulation and nitrogen homeostasis by providing iron according to internal nitrogen status
Nature Communications (2024)
-
A micro RNA mediates shoot control of root branching
Nature Communications (2023)
-
CEP peptide and cytokinin pathways converge on CEPD glutaredoxins to inhibit root growth
Nature Communications (2023)
-
Balancing nitrate acquisition strategies in symbiotic legumes
Planta (2023)
-
Root nitrate uptake in sugarcane (Saccharum spp.) is modulated by transcriptional and presumably posttranscriptional regulation of the NRT2.1/NRT3.1 transport system
Molecular Genetics and Genomics (2022)