Abstract
Plants use extracellular vesicles (EVs) to transport small RNAs (sRNAs) into their fungal pathogens and silence fungal virulence-related genes through a phenomenon called ‘cross-kingdom RNAi’. It remains unknown, however, how sRNAs are selectively loaded into EVs. Here, we identified several RNA-binding proteins in Arabidopsis, including Argonaute 1 (AGO1), RNA helicases (RHs) and annexins (ANNs), which are secreted by exosome-like EVs. AGO1, RH11 and RH37 selectively bind to EV-enriched sRNAs but not to non-EV-associated sRNAs, suggesting that they contribute to the selective loading of sRNAs into EVs. Conversely, ANN1 and ANN2 bind to sRNAs non-specifically. The ago1, rh11 rh37 and ann1 ann2 mutants showed reduced secretion of sRNAs in EVs, demonstrating that these RNA-binding proteins play an important role in sRNA loading and/or stabilization in EVs. Furthermore, rh11 rh37 and ann1 ann2 showed increased susceptibility to Botrytis cinerea, suggesting that RH11, RH37, ANN1 and ANN2 positively regulate plant immunity against B. cinerea.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available in the Supplementary Information and Source data provided with this paper or from the corresponding authors upon reasonable request. Additional data related to this study are available from the corresponding author upon request. The raw files for LC–MS/MS analyses generated during this study are available at PeptideAtlas (http://www.peptideatlas.org/PASS/PASS01572).
Change history
24 March 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41477-021-00901-5
References
Baulcombe, D. RNA silencing in plants. Nature 431, 356–363 (2004).
Mello, C. C. & Conte, D. Jr. Revealing the world of RNA interference. Nature 431, 338–342 (2004).
Huang, C. Y., Wang, H., Hu, P., Hamby, R. & Jin, H. Small RNAs—big players in plant–microbe interactions. Cell Host Microbe 26, 173–182 (2019).
Cai, Q. et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129 (2018).
Weiberg, A. et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013).
Cai, Q., He, B., Weiberg, A., Buck, A. H. & Jin, H. Small RNAs and extracellular vesicles: new mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog. 15, e1008090 (2019).
Weiberg, A. & Jin, H. Small RNAs—the secret agents in the plant–pathogen interactions. Curr. Opin. Plant Biol. 26, 87–94 (2015).
Wang, M. et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2, 16151 (2016).
Colombo, M., Raposo, G. & Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 (2014).
Kowal, J., Tkach, M. & Thery, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29, 116–125 (2014).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Lasser, C. et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 9, 9 (2011).
Luo, S. S. et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 81, 717–729 (2009).
Yan, W. et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 20, 597–609 (2018).
Kowal, J. et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl Acad. Sci. USA 113, E968–E977 (2016).
Santangelo, L. et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 17, 799–808 (2016).
Shurtleff, M. J., Temoche-Diaz, M. M., Karfilis, K. V., Ri, S. & Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5, e19276 (2016).
Hagiwara, K., Katsuda, T., Gailhouste, L., Kosaka, N. & Ochiya, T. Commitment of annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 589, 4071–4078 (2015).
Vaucheret, H. Plant ARGONAUTES. Trends Plant Sci. 13, 350–358 (2008).
Mi, S. et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008).
Zhang, X. et al. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 5, 5468 (2014).
Linder, P. & Jankowsky, E. From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 (2011).
Mingam, A. et al. DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol. J. 2, 401–415 (2004).
Rutter, B. D. & Innes, R. W. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173, 728–741 (2017).
Jeppesen, D. K. et al. Reassessment of exosome composition. Cell 177, 428–445 (2019).
Théry, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750–1535750 (2018).
Srinivasan, S. et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177, 446–462 (2019).
Thery, C., Amigorena, S., Raposo, G. & Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. 30, 3.22.1–3.22.29 (2006).
Xu, R., Greening, D. W., Rai, A., Ji, H. & Simpson, R. J. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87, 11–25 (2015).
Chow, F. W. et al. Secretion of an argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res. 47, 3594–3606 (2019).
Leidal, A. M. et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199 (2020).
Mukherjee, K. et al. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 17, 1184–1203 (2016).
Aukrust, I. et al. The mRNA-binding site of annexin A2 resides in helices C-D of its domain IV. J. Mol. Biol. 368, 1367–1378 (2007).
McKenzie, A. J. et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 15, 978–987 (2016).
Goldie, B. J. et al. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 42, 9195–9208 (2014).
Melo, S. A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014).
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004).
Burroughs, A. M. et al. Deep-sequencing of human argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin. RNA Biol. 8, 158–177 (2011).
Czech, B. & Hannon, G. J. Small RNA sorting: matchmaking for argonautes. Nat. Rev. Genet. 12, 19–31 (2011).
Arroyo, J. D. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 108, 5003–5008 (2011).
Gibbings, D. J., Ciaudo, C., Erhardt, M. & Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11, 1143–1149 (2009).
Li, S. et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574 (2013).
Akers, J. C., Gonda, D., Kim, R., Carter, B. S. & Chen, C. C. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 113, 1–11 (2013).
Wang, J. et al. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22, 4009–4030 (2010).
Wahlgren, J. et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 40, e130 (2012).
Buck, A. H. et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 5, 5488 (2014).
Wu, Z. et al. Extracellular vesicle-mediated communication within host–parasite interactions. Front. Immunol. 9, 3066 (2018).
Boavida, L. C., Qin, P., Broz, M., Becker, J. D. & McCormick, S. Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol. 163, 696–712 (2013).
Drakakaki, G. et al. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res 22, 413–424 (2012).
Vaucheret, H., Vazquez, F., Crété, P. & Bartel, D. P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197 (2004).
Acknowledgements
We thank N. Raikhel for the 35Spro::ARA6-GFP and SYP61pro::CFP-SYP61 seeds, L. Boavida for TET8pro::TET8-GFP and 35Spro::TET8-GFP seeds and R. Hamby for editing the paper. This work was supported by grants from the National Institutes of Health (R01GM093008 and R35GM136379), the National Science Foundation (IOS2017314) and AES-CE (PPA-7517H) to H.J. and the National Institutes of Health (R01 ES025121) to Y.W. Q.C. is supported by funds from National Natural Science Foundation of China (32070288).
Author information
Authors and Affiliations
Contributions
H.J. conceived the idea and supervised the project. B.H., Q.C. and H.J. designed the experiments. B.H. and Q.C. performed most of the experiments and analysed data. L.Q. and T.H. contributed to the functional analysis of the EV-associated RBPs. S.W. generated the ANN2-CFP line and C.H. generated the rh11 rh37 double mutant. W.M. and Y.W. performed mass spectrometry and conducted bioinformatics analysis. B.H., Q.C. and H.J. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Michael Feldbrügge and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Images show the various steps in plant EVs isolation.
a, Leaves were harvested and rinsed; the petioles were removed. Next, leaves were placed in a syringe with infiltration buffer and vacuumed. b, The leaves were taped in the same orientation onto a 1 ml syringe after infiltration buffer was vacuumed into leaves. c, Syringe with taped leaves was placed into a 50 ml conical tube. d, The apoplastic wash was collected by gently centrifuging the leaves at 900 g at 4 °C. e, Scheme of EV isolation by differential ultracentrifugation from apoplastic wash of Arabidopsis. Sup, Supernatant. f, Confocal microscopy images of P40 and P100 fractions isolation by ultracentrifugation from apoplastic wash of TET8pro:TET8-GFP plants. Equivalent amounts of plants were inoculated with B. cinerea for 36 hours before P40 and P100 fraction isolation. Scale bars, 10 μm. g, GFP-labelled TET8 was detectable in both P40 and P100 EV fractions by western blot. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 2 Gene ontology (GO) enrichment analysis of proteins enriched in EVs.
a, b, The plant EV proteome from B. cinerea-infected Arabidopsis plants was categorized based on GO terms related to biological process (a) and molecular function (b) through The Arabidopsis Information Resource Web site (www.arabidopsis.org).
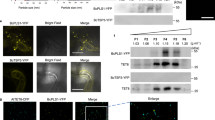
Extended Data Fig. 3 TET8 and PEN1 localization.
a, Confocal microscopy images of TET8-YFP and ARA6-CFP at B. cinerea infection site on N. benthamiana. TET8-YFP was partially colocalized with ARA6-CFP signals. b, Fluorescent intensity was quantified for the images used in (a). Transections used for fluorescence intensity measurements are indicated by blue lines. Green and red lines represent histograms of ARA6-CFP and TET8-YFP fluorescent intensities, respectively. c, Confocal microscopy images of CFP-PEN1 and ARA6-YFP at the B. cinerea infection site on N. benthamiana. CFP-PEN1 did not colocalize with ARA6-YFP signals. d, Fluorescence intensity was quantified for the images used in (c). Transections used for fluorescence intensity measurements are indicated by blue lines. Green and red lines represent histograms of CFP-PEN1 and ARA6-YFP fluorescent intensities, respectively. Scale bars, 10 μm.
Extended Data Fig. 4 Bottom-loading EV separation by sucrose gradient centrifugation.
a, Pellets obtained from 100,000 g centrifugations (P100) were used to perform sucrose gradient centrifugation by both top and bottom loading. b, Six fractions were collected from bottom-loading plant EV sucrose gradient centrifugation. TET8, AGO1, RH11, RH37, ANN1, ANN2 were detected by western blot. EV-enriched (TAS1c-siR483, TAS2-siR453 and miR166) and non-EV-associated (TAS1c-siR585 and TAS2-siR710) sRNAs were detected by RT–PCR. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 5 Colocalization between EV-associated RBPs with MVB marker ARA6 and EV marker TET8.
a, b, Fluorescent protein-labelled RBPs were co-expressed transiently with MVB marker ARA6 (a) and EV marker TET8 (b) in N. benthamiana. Confocal microscopy was used to determine the localization of RBPs (AGO1, RH11, RH37, ANN1, ANN2) with ARA6 and TET8. AGO2 was used as a control. Scale bars, 10 μm. The experiments were repeated three times independently with similar results.
Extended Data Fig. 6 sRNAs specifically bound by AGO2, AGO4 and AGO5 were absent from plant EVs.
AGO2-associated miR393*, AGO4-associated siR1003, AGO5-associated miR390* and both AGO1 and AGO5-associated miR156 were examined in isolated plant EVs by sRNA RT–PCR. TAS1c-siR483, TAS2-siR453 and miR166 were detected in EVs and used as positive controls. TAS1c-siR585 and TAS2-siR710 were used as negative controls. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 7 AGO1 and RH37 selectively bind EV-enriched sRNAs in N. benthamiana.
YFP-AGO1 or RH37-CFP were co-expressed with EV-enriched (TAS1c-siR483, TAS2-siR453 and miR166) and non-EV-associated sRNAs (TAS1c-siR585 and TAS2-siR710) in N. benthamiana, sRNAs were immunoprecipitated from plant total extraction using antibodies against GFP and detected by sRNA RT–PCR. IgG was used as a negative control. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 8 Verification of rh11rh37 and ann1ann2 double mutants.
a, The developmental phenotypes of the rh11rh37 double mutant that both RH11 and RH37 expression was suppressed by artificial miRNA in Col-0. b, The real-time RT–PCR analysis of RH11 and RH37 expression in rh11rh37 mutants. The data are presented as mean ± s.d., n = 3 biologically independent replicates. The error bars indicate the standard deviation (s.d.). c, The developmental phenotypes of the rh11rh37#6 mutant that RH37 expression was suppressed by artificial miRNA in rh11 knockout mutant. d, RT–PCR analysis of RH11 and real-time PCR analysis of RH37 in rh11rh37#6 mutant. The data are presented as mean ± s.d., n = 3 biologically independent replicates. The error bars indicate the standard deviation (s.d.). e, Phenotype of wild-type and ann1ann2 mutant grown for 4 weeks in a growth chamber. f, RT–PCR analysis of the expression levels of ANN1 and ANN2 in wild-type and ann1ann2 double mutant. The statistical analysis in b was performed using ANOVA Dunnett’s multiple comparisons test. The statistical analysis in d was performed using unpaired two-tailed Student’s t-tests. The small open circles represent the individual values. The asterisks indicate significant differences: *P < 0.05, **P < 0.01, ***P < 0.001. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 9 EV-enriched sRNA amount was reduced in EVs isolated from the mutants of EV-associated RBPs.
The relative level of both EV-enriched and non-EV-associated sRNAs were examined by quantitative real-time RT–PCR in the total fraction and EV fraction from ago1-27 (a), rh11rh37 (b) and ann1ann2 (c) mutants. The data are presented as mean ± s.d., n = 3 biologically independent replicates. The error bars indicate the standard deviation (s.d.). The statistical analysis was performed using unpaired two-tailed Student’s t-tests. The small open circles represent the individual values. The asterisks indicate significant differences: **P < 0.01, ***P < 0.01, NS, not significant.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots and gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots and gels.
Source Data Fig. 5
Unprocessed gels.
Source Data Fig. 6
Unprocessed western blots and gels.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots and gels.
Source Data Extended Data Fig. 6
Unprocessed gels.
Source Data Extended Data Fig. 7
Unprocessed western blots and gels.
Source Data Extended Data Fig. 8
Unprocessed gels.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
He, B., Cai, Q., Qiao, L. et al. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 7, 342–352 (2021). https://doi.org/10.1038/s41477-021-00863-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00863-8
This article is cited by
-
Leafhopper salivary vitellogenin mediates virus transmission to plant phloem
Nature Communications (2024)
-
Molecular mechanisms and therapeutic application of extracellular vesicles from plants
Molecular Biology Reports (2024)
-
Frontiers in plant RNA research in ICAR2023: from lab to innovative agriculture
Plant Molecular Biology (2024)
-
TCTP regulates genotoxic stress and tumorigenicity via intercellular vesicular signaling
EMBO Reports (2024)
-
Nanoparticle carriers enhance RNA stability and uptake efficiency and prolong the protection against Rhizoctonia solani
Phytopathology Research (2023)