Abstract
Root-associated soil bacteria can strongly influence plant fitness. DNA methylation is an epigenetic mark important to many fundamental biological processes; however, its roles in plant interactions with beneficial microbes remain elusive. Here, we report that active DNA demethylation in Arabidopsis controls root secretion of myo-inositol and consequently plant growth promotion triggered by Bacillus megaterium strain YC4. Root-secreted myo-inositol is critical for YC4 colonization and preferentially attracts B. megaterium among the examined bacteria species. Active DNA demethylation antagonizes RNA-directed DNA methylation in controlling myo-inositol homeostasis. Importantly, we demonstrate that active DNA demethylation controls myo-inositol-mediated mutualism between YC4 and Solanum lycopersicum, thus suggesting a conserved nature of this epigenetic regulatory mechanism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The mRNA-Seq and WGBS data from this publication have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Gene Expression Omnibus Series accession number GSE83802 (ref. 23). Source data are provided with this paper.
References
Lugtenberg, L. & Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 (2009).
Nobori, T., Mine, A. & Tsuda, K. Molecular networks in plant–pathogen holobiont. FEBS Lett. 592, 1937–1953 (2018).
Sasse, J., Martinoia, E. & Northen, T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41 (2018).
Zhang, H., Lang, Z. & Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506 (2018).
Lang, Z. et al. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl Acad. Sci. USA 114, E4511–E4519 (2017).
Liu, R. et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl Acad. Sci. USA 112, 10804–10809 (2015).
Zhu, J.-K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166 (2009).
Le, T.-N. et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 15, 458 (2014).
Qian, W. et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336, 1445–1448 (2012).
Vilchez, J. I. et al. Genome sequence of Bacillus megaterium strain YC4-R4, a plant growth-promoting rhizobacterium isolated from a high-salinity environment. Genome Announc. 6, e00527-18 (2018).
Onodera, Y. et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622 (2005).
Wierzbicki, A. T., Haag, J. R. & Pikaard, C. S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648 (2008).
Pini, F. et al. Bacterial biosensors for in vivo spatiotemporal mapping of root secretion. Plant Physiol. 174, 1289–1306 (2017).
Sannino, A., Demitri, C. & Madaghiele, M. Biodegradable cellulose-based hydrogels: design and applications. Materials 2, 353–373 (2009).
Torabinejad, J., Donahue, J. L., Gunesekera, B. N., Allen-Daniels, M. J. & Gillaspy, G. E. VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 150, 951–961 (2009).
Morcillo, R. J. et al. Rhizobacterium-derived diacetyl modulates plant immunity in a phosphate-dependent manner. EMBO J. 39, e102602 (2019).
Vilchez, J. I. et al. Complete genome sequence of Bacillus megaterium strain TG1-E1, a plant drought tolerance-enhancing bacterium. Microbiol. Resour. Announc. 7, e00842-18 (2018).
Bais, H. P., Fall, R. & Vivanco, J. M. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134, 307–319 (2004).
Yu, A. et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl Acad. Sci. USA 110, 2389–2394 (2013).
López Sánchez, A., Stassen, J. H., Furci, L., Smith, L. M. & Ton, J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. Cell Mol. Biol. 88, 361–374 (2016).
Ma, L. et al. Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant 9, 541–557 (2016).
Glawischnig, E. Camalexin. Phytochemistry 68, 401–406 (2007).
Tang, K., Lang, Z., Zhang, H. & Zhu, J.-K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2, 16169 (2016).
Bacete, L., Mélida, H., Miedes, E. & Molina, A. Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. Cell Mol. Biol. 93, 614–636 (2018).
Schulze-Lefert, P. Knocking on the heaven’s wall: pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr. Opin. Plant Biol. 7, 377–383 (2004).
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren van Themaat, E. & Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838 (2013).
Busby, P. E. et al. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15, e2001793 (2017).
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Salas González, I. & Dangl, J. L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163 (2017).
Martin, F. M., Uroz, S. & Barker, D. G. Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356, eaad4501 (2017).
Gillaspy, G. E. The cellular language of myo-inositol signaling. New Phytol. 192, 823–839 (2011).
Yoshida, K. I., Aoyama, D., Ishio, I., Shibayama, T. & Fujita, Y. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179, 4591–4598 (1997).
Vives-Peris, V., de Ollas, C, Gómez-Cadenas, A. & Pérez-Clemente, R. M. Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17 (2020).
Lung, S. C. et al. Phytase activity in tobacco (Nicotiana tabacum) root exudates is exhibited by a purple acid phosphatase. Phytochemistry 69, 365–373 (2008).
Jones, P., Garcia, B. J., Furches, A., Tuskan, G. A. & Jacobson, D. Plant host-associated mechanisms for microbial selection. Front. Plant Sci. 10, 862 (2019).
Kohler, P. R. A., Zheng, J. Y., Schoffers, E. & Rossbach, S. Inositol catabolism, a key pathway in Sinorhizobium meliloti for competitive host nodulation. Appl. Environ. Microbiol. 76, 7972–7980 (2010).
Donahue, J. L. et al. The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 22, 888–903 (2010).
Kanter, U. et al. The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221, 243–254 (2005).
Lei, M. et al. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl Acad. Sci. USA 112, 3553–3557 (2015).
Williams, B. P., Pignatta, D., Henikoff, S. & Gehring, M. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 11, e1005142 (2015).
Kawakatsu, T. et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166, 492–505 (2016).
Satgé, C. et al. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat. Plants 2, 16166 (2016).
Huang, A. C. et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364, eaau6389 (2019).
Stringlis, I. A. et al. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl Acad. Sci. USA 115, E5213–E5222 (2018).
Voges, M. J. E. E. E., Bai, Y., Schulze-Lefert, P. & Sattely, E. S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl Acad. Sci. USA 116, 12558–12565 (2019).
Hamon, M. A. & Lazazzera, B. A. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42, 1199–1209 (2001).
López, A. & Alippi, A. Phenotypic and genotypic diversity of Bacillus cereus isolates recovered from honey. Int. J. Food Microbiol. 117, 175–184 (2007).
Heras, J., Domínguez, C., Mata, E. & Pascual, V. GelJ—a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 16, 270 (2015).
Hu, L. et al. Root exudate metabolites drive plant–soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738 (2018).
Walker, T. S., Bais, H. P., Grotewold, E. & Vivanco, J. M. Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51 (2003).
Roessner, U., Wagner, C., Kopka, J., Trethewey, R. N. & Willmitzer, L. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography–mass spectrometry. Plant J. Cell Mol. Biol. 23, 131–142 (2000).
Barsch, A., Carvalho, H. G., Cullimore, V. J. & Niehaus, K. GC–MS based metabolite profiling implies three interdependent ways of ammonium assimilation in Medicago truncatula root nodules. J. Biotechnol. 127, 79–83 (2006).
Gorzolka, K., Lissel, M., Kessler, N., Loch-Ahring, S. & Niehaus, K. Metabolite fingerprinting of barley whole seeds, endosperms, and embryos during industrial malting. J. Biotechnol. 159, 177–187 (2012).
Kopka, J. et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21, 1635–1638 (2005).
Ge, S. X., Son, E. W. & Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics 19, 534 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Xi, Y. & Li, W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009).
Cox, M. P., Peterson, D. A. & Biggs, P. J. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11, 485 (2010).
Juhling, F. et al. Metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 26, 256–262 (2016).
Acknowledgements
We thank the core facilities of Genomics, Plant Proteomics and Metabolomics, and Plant Cell Biology at the Shanghai Center for Plant Stress Biology for sequencing, GC–MS and microscopic analyses, respectively. We also express special thanks to A. Álvarez for artistic collaboration in the design of the model presented in this manuscript. Research in the laboratory of H.Z. has been supported by the Chinese Academy of Sciences and Thousand Talents Program for Young Scientists, China. J.I.V. was supported by the Chinese Academy of Sciences President’s International Fellowship Initiative fellowship.
Author information
Authors and Affiliations
Contributions
H. Zhang designed the project. J.I.V. performed or led all of the experiments and coordinated with Y.Y. for the RNA-Seq and WGBS data analyses. H.Zi, L.P., R.L. and K.T. analysed RNA-Seq and/or WGBS raw data for the DEG and DMR lists. D.H., S.L., R.K., W.W., W.H., Z.L. and D.M. participated in the experiments and/or data analyses. H. Zhang and J.I.V. wrote the manuscript with input from P.W.P., C.-P.S. and J.-K.Z.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Glenda Gillaspy, Mingbo Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
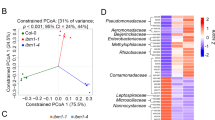
Extended Data Fig. 1 Bacillus megaterium YC4-R4 induces growth-promotion in various plant species (Related to Fig. 1).
a, Arabidopsis thaliana and Brachypodium distachyon showed YC4-R4-induced growth-promotion. Plant images are of the same scale, as indicated by the vertical rulers. These experiments were repeated three times with similar results (n = 6 biological independent samples). b–d, YC4-R4 induced plant growth-promotion in Solanum lycopersicum, Brassica napus and Capsicum annuum, respectively. The lower panels show quantification of whole plant or shoots dry weight. The boxplots and the pictures show representative data from three independent experiments (Solanum lycopersicum and Capsicum annuum, n = 18; Brassica napus, n = 16 biological independent samples, respectively). Whiskers represent the min to max data range; the median is represented by the central horizontal line. The upper and lower limits of the box outline represent the first and third quartile. All sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences. e, YC4-R4 induced plant growth-promotion in Arabidopsis mutants (nrpd1-3 and nrpe1-11) with defective RNA-directed DNA methylation, but not in the rdd mutant that is defective in DNA demethylation. Plants were pictured at 14 DAT (left panel) and their dry weights were quantified (right panel). The boxplots show representative data from three independent experiments (n = 30). Whiskers represent the min to max data range; the median is represented by the central horizontal line. The upper and lower limits of the box outline represent the first and third quartile. All sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences. In all data sets described above, error bar means SD.
Extended Data Fig. 2 myo-Inositol supplementation to the Arabidopsis DNA demethylation mutants restored root association with B. megaterium YC4-R4 (Related to Fig. 2).
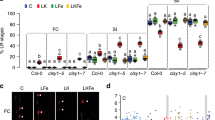
a, Quantification of root-associated bacteria from plants that were grown in sterile growth medium, treated with YC4-R4 and colonization rate was recorded at 14 DAT. The boxplots show representative data from three independent experiments (n = 30 biological independent samples). Whiskers represent the min to max data range, the median is represented by the central horizontal line. The upper and lower limits of the box outline represent the first and third quartile. Sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences. In all data sets described above, error bar means SD. b, Visualization of root-associated bacteria from plants that were grown in sterile growth medium and were treated with YC4-R4 at 5 DAT. The green arrows point to example spots where bacteria are present. This experiment was carried out three times with similar results (n = 5 biological independent samples).
Extended Data Fig. 3 myo-Inositol restored B. megaterium YC4-R4 biofilm formation on the roots of rdd and ros1 (Related to Fig. 4).
Roots of Col-0, rdd and ros1 were shown in bright field images (a) and Scanning Electronic Microscopy (SEM) images (b). For samples with no visualized biofilm, less amplified images were used to show no visual findings. Red area or arrows indicate bacteria in the bright field images. Blue area indicates biofilm in the SEM images. In both cases, images show representative pictures from three independent experiments with similar results (n = 5 biological independent samples).
Extended Data Fig. 4 myo-Inositol preferentially influences Bacillus megaterium (Related to Fig. 4).
a, Root colonization of the bacteria including B. megaterium YC4-R4, B. megaterium TG1-E1, B. amyloliquefaciens GB03 and P. syringe DC3000. The effects of myo-inositol on bacteria colonization to rdd roots were statistically analyzed based on each bacteria strain. The bar graph shows representative data from three independent experiments (n = 3 biological independent samples). Sets of data were analyzed by one-way ANOVA including a Tukey’s test, where * means statistical difference by p < 0.05. b, Gene Ontology (GO) categorization of DEGs that were up-regulated in myo-inositol-treated YC4-R4. c, GO categorization of DEGs that were down-regulated in myo-inositol-treated YC4-R4. d, Relative expression levels of a group of key genes involved in bacteria colonization-related activities. The bar graphs show representative quantitative real-time PCR results from three independent experiments (n = 3 biological independent samples). Sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences with mock set. In all data sets described above, error bar means SD.
Extended Data Fig. 5 Transcriptional regulation of plant responses to YC4-R4 and myo-inositol homeostasis are altered by defective DNA demethylation (Related to Fig. 5).
a, Gene Ontology (GO) categorization of YC4-R4-induced DEGs in rdd. Diagrams are designed based on Cytoscape Software. The size of solid-lined circles represents the number of genes in each GO category. Scale color bar indicates the significance of gene expression in each GO category. Clustered biological processes were indicated manually by dash-lined gray circles, and the GO term is underlined if the cluster is present only in either the YC4-R4-induced or YC-repressed DEGs. b, GO categorization of YC4-R4-repressed DEGs in rdd. c, Relative expression levels of myo-inositol homeostasis genes in Col-0, rdd and ros1. The bar graph shows representative quantitative real-time PCR results from three independent experiments (n = 3 biological independent samples). Sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences with Col-0 set. In all data sets described above, error bar means SD.
Extended Data Fig. 6 ROS1 counteracts RdDM in controlling DNA methylation at gene-associated loci (Related to Fig. 6).
a, Snapshots of WGBS results that show DNA hyper methylation at the vicinity regions of myo-inositol homeostasis genes in rdd with (T) and without (M) YC4-R4 treatment. b, Snapshots of WGBS results showing that ROS1-mediated DNA demethylation counteracts RdDM at the vicinity regions of myo-inositol homeostasis genes. c, Categorization of Arabidopsis genomic loci where DNA methylation is co-regulated by ROS1 and RdDM. Gene-associated regions were further subject to GO analysis, revealing an enrichment of genes related to cell wall organization.
Extended Data Fig. 7 Tomato myo-inositol homeostasis genes are subject to the regulation by SlDML2-dependent DNA demethylation (Related to Fig. 7).
a, Quantification of plant dry weights at 14 DAT. The boxplots show representative data from three independent experiments (n = 18 biological independent samples). Whiskers represent the min to max data range; the median is represented by the central horizontal line. The upper and lower limits of the box outline represent the first and third quartile. In both cases (C and D), sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences. b, Relative expression levels of three tomato homologs of Arabidopsis myo-inositol homeostasis genes. The bar graphs show representative quantitative real-time PCR results from three independent experiments (n = 3 biological independent samples). Sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences with WT set. c, Snapshots of WGBS results that show DNA hyper methylation associated with myo-ionsitol homeostasis genes in tomato. d, A heat map of myo-inositol catabolism DEGs identified by RNAseq in myo-inositol-treated YC4-R4 compared to YC4-R4 under the mock condition. Values are representatives from three independent experiments (n = 4 biological independent samples). e, Relative expression levels of myo-inositol catabolism genes in YC4. The bar graph shows representative quantitative real-time PCR results from three independent experiments (n = 3 biological independent samples). Sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences with mock set. f, The effects of L-threonin on YC4-R4 chemotaxis responses. The bar graph shows representative RFU (relative fluorescent unit) values from three independent experiments (n = 9 biological independent samples). Sets of data were compared by t-student test by two-tailed way and 95% confident intervals (p < 0.05), in which * means statistical differences with mock set. In all data sets described above, error bar means SD.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables
Supplementary Tables 1–3.
Supplementary Data
Statistical values relating to Figs. 1–7.
Computational Data
Statistical values relating to Extended Data Figs. 1–7 and Supplementary Fig. 1.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 7
Statistical Source Data.
Rights and permissions
About this article
Cite this article
Vílchez, J.I., Yang, Y., He, D. et al. DNA demethylases are required for myo-inositol-mediated mutualism between plants and beneficial rhizobacteria. Nat. Plants 6, 983–995 (2020). https://doi.org/10.1038/s41477-020-0707-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0707-2
This article is cited by
-
A volatile producing Bacillus subtilis strain from the rhizosphere of Haloxylon ammodendron promotes plant root development
Plant and Soil (2023)
-
Long-term effect of epigenetic modification in plant–microbe interactions: modification of DNA methylation induced by plant growth-promoting bacteria mediates promotion process
Microbiome (2022)
-
Dysfunction of histone demethylase IBM1 in Arabidopsis causes autoimmunity and reshapes the root microbiome
The ISME Journal (2022)
-
Abiotic stress responses in plants
Nature Reviews Genetics (2022)
-
Flavonoid-attracted Aeromonas sp. from the Arabidopsis root microbiome enhances plant dehydration resistance
The ISME Journal (2022)