Abstract
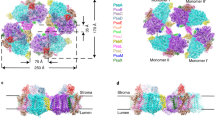
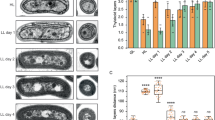
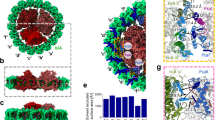
Cyanobacterial thylakoid membranes represent the active sites for both photosynthetic and respiratory electron transport. We used high-resolution atomic force microscopy to visualize the native organization and interactions of photosynthetic complexes within the thylakoid membranes from the model cyanobacterium Synechococcus elongatus PCC 7942. The thylakoid membranes are heterogeneous and assemble photosynthetic complexes into functional domains to enhance their coordination and regulation. Under high light, the chlorophyll-binding proteins IsiA are strongly expressed and associate with Photosystem I (PSI), forming highly variable IsiA−PSI supercomplexes to increase the absorption cross-section of PSI. There are also tight interactions of PSI with Photosystem II (PSII), cytochrome b6f, ATP synthase and NAD(P)H dehydrogenase complexes. The organizational variability of these photosynthetic supercomplexes permits efficient linear and cyclic electron transport as well as bioenergetic regulation. Understanding the organizational landscape and environmental adaptation of cyanobacterial thylakoid membranes may help inform strategies for engineering efficient photosynthetic systems and photo-biofactories.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Liu, L. N. Distribution and dynamics of electron transport complexes in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta 1857, 256–265 (2016).
Vermaas, W. F. in Encyclopedia of Life Sciences 245–251 (Nature Publishing Group, 2001).
Mullineaux, C. W. Co-existence of photosynthetic and respiratory activities in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta 1837, 503–511 (2014).
Saer, R. G. & Blankenship, R. E. Light harvesting in phototrophic bacteria: structure and function. Biochem. J. 474, 2107–2131 (2017).
Chen, H.-Y. S., Bandyopadhyay, A. & Pakrasi, H. B. Function, regulation and distribution of IsiA, a membrane-bound chlorophyll a-antenna protein in cyanobacteria. Photosynthetica 56, 322–333 (2018).
Bibby, T. S., Nield, J. & Barber, J. Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743–745 (2001).
Boekema, E. J. et al. A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412, 745–748 (2001).
Vinnemeier, J., Kunert, A. & Hagemann, M. Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 169, 323–330 (1998).
Havaux, M. et al. The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Lett. 579, 2289–2293 (2005).
Yousef, N., Pistorius, E. K. & Michel, K. P. Comparative analysis of idiA and isiA transcription under iron starvation and oxidative stress in Synechococcus elongatus PCC 7942 wild-type and selected mutants. Arch. Microbiol. 180, 471–483 (2003).
Toporik, H., Li, J., Williams, D., Chiu, P. L. & Mazor, Y. The structure of the stress-induced photosystem I-IsiA antenna supercomplex. Nat. Struct. Mol. Biol. 26, 443–449 (2019).
Cao, P. et al. Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex. Nat. Plants 6, 167–176 (2020).
Ma, F. et al. Dynamic changes of IsiA-containing complexes during long-term iron deficiency in Synechocystis sp PCC 6803. Mol. Plant 10, 143–154 (2017).
Sun, J. L. & Golbeck, J. H. The presence of the IsiA–PSI supercomplex leads to enhanced photosystem I electron throughput in iron-starved cells of Synechococcus sp. PCC 7002. J. Phys. Chem. B 119, 13549–13559 (2015).
Chauhan, D. et al. A novel photosynthetic strategy for adaptation to low-iron aquatic environments. Biochemistry 50, 686–692 (2011).
Park, Y. I., Sandstrom, S., Gustafsson, P. & Oquist, G. Expression of the isiA gene is essential for the survival of the cyanobacterium Synechococcus sp. PCC 7942 by protecting photosystem II from excess light under iron limitation. Mol. Microbiol 32, 123–129 (1999).
Schoffman, H. & Keren, N. Function of the IsiA pigment-protein complex in vivo. Photosynth. Res. 141, 343–353 (2019).
Busch, K. B., Deckers-Hebestreit, G., Hanke, G. T. & Mulkidjanian, A. Y. Dynamics of bioenergetic microcompartments. Biol. Chem. 394, 163–188 (2013).
Casella, S. et al. Dissecting the native architecture and dynamics of cyanobacterial photosynthetic machinery. Mol. Plant 10, 1434–1448 (2017).
MacGregor-Chatwin, C. et al. Lateral segregation of photosystem I in cyanobacterial thylakoids. Plant Cell 29, 1119–1136 (2017).
Bečková, M. et al. Association of Psb28 and Psb27 proteins with PSII-PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol. Plant 10, 62–72 (2017).
Liu, H. et al. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 342, 1104–1107 (2013).
Liu, L. N. & Scheuring, S. Investigation of photosynthetic membrane structure using atomic force microscopy. Trends Plant Sci. 18, 277–286 (2013).
MacGregor-Chatwin, C. et al. Membrane organization of photosystem I complexes in the most abundant phototroph on Earth. Nat. Plants 5, 879–889 (2019).
Riediger, M. et al. Biocomputational analyses and experimental validation identify the regulon controlled by the redox-responsive transcription factor RpaB. iScience 15, 316–331 (2019).
Kappell, A. D., Bhaya, D. & van Waasbergen, L. G. Negative control of the high light-inducible hliA gene and implications for the activities of the NblS sensor kinase in the cyanobacterium Synechococcus elongatus strain PCC 7942. Arch. Microbiol. 186, 403–413 (2006).
Liu, L. N. et al. Control of electron transport routes through redox-regulated redistribution of respiratory complexes. Proc. Natl Acad. Sci. USA 109, 11431–11436 (2012).
Sun, Y. et al. Light modulates the biosynthesis and organization of cyanobacterial carbon fixation machinery through photosynthetic electron flow. Plant Physiol. 171, 530–541 (2016).
Sun, Y., Wollman, A. J. M., Huang, F., Leake, M. C. & Liu, L. N. Single-organelle quantification reveals the stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell 31, 1648–1664 (2019).
Jordan, P. et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917 (2001).
Yeremenko, N. et al. Supramolecular organization and dual function of the IsiA chlorophyll-binding protein in cyanobacteria. Biochemistry 43, 10308–10313 (2004).
Zipfel, W. & Owens, T. G. Calculation of absolute photosystem I absorption cross-sections from P700 photo-oxidation kinetics. Photosynth. Res. 29, 23–35 (1991).
Ryan-Keogh, T. J., Macey, A. I., Cockshutt, A. M., Moore, C. M. & Bibby, T. S. The cyanobacterial chlorophyll-binding-protein Isia acts to increase the in vivo effective absorption cross-section of PSI under iron limitation. J. Phycol. 48, 145–154 (2012).
Odom, W. R., Hodges, R., Chitnis, P. R. & Guikema, J. A. Characterization of Synechocystis sp. PCC 6803 in iron-supplied and iron-deficient media. Plant Mol. Biol. 23, 1255–1264 (1993).
Yadav, K. N. et al. Supercomplexes of plant photosystem I with cytochrome b6f, light-harvesting complex II and NDH. Biochim. Biophys. Acta Bioenerg. 1858, 12–20 (2017).
Steinbeck, J. et al. Structure of a PSI-LHCI-cyt b6f supercomplex in Chlamydomonas reinhardtii promoting cyclic electron flow under anaerobic conditions. Proc. Natl Acad. Sci. USA 115, 10517–10522 (2018).
Rast, A. et al. Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nat. Plants 5, 436–446 (2019).
Hellmich, J. et al. Native-like photosystem II superstructure at 2.44 A resolution through detergent extraction from the protein crystal. Structure 22, 1607–1615 (2014).
Umena, Y., Kawakami, K., Shen, J. R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A. Nature 473, 55–60 (2011).
Chang, L. et al. Structural organization of an intact phycobilisome and its association with photosystem II. Cell Res. 25, 726–737 (2015).
Zlenko, D. V., Galochkina, T. V., Krasilnikov, P. M. & Stadnichuk, I. N. Coupled rows of PBS cores and PSII dimers in cyanobacteria: symmetry and structure. Photosynth. Res. 133, 245–260 (2017).
McConnell, M. D., Koop, R., Vasil’ev, S. & Bruce, D. Regulation of the distribution of chlorophyll and phycobilin-absorbed excitation energy in cyanobacteria. A structure-based model for the light state transition. Plant Physiol. 130, 1201–1212 (2002).
Seelert, H., Dencher, N. A. & Müller, D. J. Fourteen protomers compose the oligomer III of the proton-rotor in spinach chloroplast ATP synthase. J. Mol. Biol. 333, 337–344 (2003).
Hahn, A., Vonck, J., Mills, D. J., Meier, T. & Kuhlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360, eaat4318 (2018).
Pogoryelov, D. et al. The oligomeric state of c rings from cyanobacterial F-ATP synthases varies from 13 to 15. J. Bacteriol. 189, 5895–5902 (2007).
Strauss, M., Hofhaus, G., Schroder, R. R. & Kuhlbrandt, W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27, 1154–1160 (2008).
Daum, B., Nicastro, D., Il, J. A., McIntosh, J. R. & Kuhlbrandt, W. Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22, 1299–1312 (2010).
Schuller, J. M. et al. Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 363, 257–260 (2019).
Laughlin, T. G., Bayne, A. N., Trempe, J. F., Savage, D. F. & Davies, K. M. Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566, 411–414 (2019).
Arteni, A. A. et al. Structural characterization of NDH-1 complexes of Thermosynechococcus elongatus by single particle electron microscopy. Biochim. Biophys. Acta 1757, 1469–1475 (2006).
Folea, I. M. et al. Single particle analysis of thylakoid proteins from Thermosynechococcus elongatus and Synechocystis 6803: localization of the CupA subunit of NDH-1. FEBS Lett. 582, 249–254 (2008).
Birungi, M. et al. Possibilities of subunit localization with fluorescent protein tags and electron microscopy examplified by a cyanobacterial NDH-1 study. Biochim. Biophys. Acta 1797, 1681–1686 (2010).
Peltier, G., Aro, E. M. & Shikanai, T. NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu. Rev. Plant Biol. 67, 55–80 (2016).
Schuller, J. M. et al. Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I. Nat. Commun. 11, 494 (2020).
Pan, X. et al. Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase. Nat. Commun. 11, 610 (2020).
Zhang, C. et al. Structural insights into NDH-1 mediated cyclic electron transfer. Nat. Commun. 11, 888–888 (2020).
Kouril, R. et al. Structural characterization of a plant photosystem I and NAD(P)H dehydrogenase supercomplex. Plant J. 77, 568–576 (2014).
Peng, L., Fukao, Y., Fujiwara, M., Takami, T. & Shikanai, T. Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21, 3623–3640 (2009).
Gao, F. et al. The NDH–1L-PSI supercomplex is important for efficient cyclic electron transport in cyanobacteria. Plant Physiol. 172, 1451–1464 (2016).
Singh, A. K. & Sherman, L. A. Iron-independent dynamics of IsiA production during the transition to stationary phase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 256, 159–164 (2006).
Bibby, T. S., Nield, J. & Barber, J. Three-dimensional model and characterization of the iron stress-induced CP43’-photosystem I supercomplex isolated from the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 276, 43246–43252 (2001).
Kouril, R. et al. Photosystem I trimers from Synechocystis PCC 6803 lacking the PsaF and PsaJ subunits bind an IsiA ring of 17 units. Biochim. Biophys. Acta 1607, 1–4 (2003).
Aspinwall, C. L., Duncan, J., Bibby, T., Mullineaux, C. W. & Barber, J. The trimeric organisation of photosystem I is not necessary for the iron-stress induced CP43' protein to functionally associate with this reaction centre. FEBS Lett. 574, 126–130 (2004).
Kouril, R. et al. Supercomplexes of IsiA and photosystem I in a mutant lacking subunit PsaL. Biochim. Biophys. Acta 1706, 262–266 (2005).
Sarcina, M. & Mullineaux, C. W. Mobility of the IsiA chlorophyll-binding protein in cyanobacterial thylakoid membranes. J. Biol. Chem. 279, 36514–36518 (2004).
Riethman, H. C. & Sherman, L. A. Purification and characterization of an iron stress-induced chlorophyll-protein from the cyanobacterium Anacystis nidulans R2. Biochim. Biophys. Acta 935, 141–151 (1988).
Burnap, R. L., Troyan, T. & Sherman, L. A. The highly abundant chlorophyll-protein complex of iron-deficient Synechococcus sp. PCC7942 (CP43') is encoded by the isiA gene. Plant Physiol. 103, 893–902 (1993).
Sandstrom, S., Park, Y. I., Oquist, G. & Gustafsson, P. CP43’, the isiA gene product, functions as an excitation energy dissipator in the cyanobacterium Synechococcus sp. PCC 7942. Photochem. Photobiol. 74, 431–437 (2001).
Zhang, J. et al. Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 551, 57–63 (2017).
Liu, L. N., Chen, X. L., Zhang, Y. Z. & Zhou, B. C. Characterization, structure and function of linker polypeptides in phycobilisomes of cyanobacteria and red algae: An overview. Biochim. Biophys. Acta Bioenerg. 1708, 133–142 (2005).
Arteni, A. A. et al. Structure and organization of phycobilisomes on membranes of the red alga Porphyridium cruentum. Photosynth. Res. 95, 169–174 (2008).
Liu, L. N. et al. Light-induced energetic decoupling as a mechanism for phycobilisome-related energy dissipation in red algae: a single molecule study. PLoS ONE 3, e3134 (2008).
Zhao, L. S. et al. Supramolecular architecture of photosynthetic membrane in red algae in response to nitrogen starvation. Biochim. Biophys. Acta 1857, 1751–1758 (2016).
Green, B. R. What happened to the phycobilisome? Biomolecules 9, 748 (2019).
Straskova, A. et al. Pigment-protein complexes are organized into stable microdomains in cyanobacterial thylakoids. Biochim. Biophys. Acta Bioenerg. 1860, 148053 (2019).
Folea, I. M., Zhang, P., Aro, E. M. & Boekema, E. J. Domain organization of photosystem II in membranes of the cyanobacterium Synechocystis PCC6803 investigated by electron microscopy. FEBS Lett. 582, 1749–1754 (2008).
Liu, L. N. et al. Watching the native supramolecular architecture of photosynthetic membrane in red algae: topography of phycobilisomes and their crowding, diverse distribution patterns. J. Biol. Chem. 283, 34946–34953 (2008).
Lenn, T., Leake, M. C. & Mullineaux, C. W. Clustering and dynamics of cytochrome bd-I complexes in the Escherichia coli plasma membrane in vivo. Mol. Microbiol. 70, 1397–1407 (2008).
Llorente-Garcia, I. et al. Single-molecule in vivo imaging of bacterial respiratory complexes indicates delocalized oxidative phosphorylation. Biochim. Biophys. Acta 1837, 811–824 (2014).
Johnson, A. S., van Horck, S. & Lewis, P. J. Dynamic localization of membrane proteins in Bacillus subtilis. Microbiology 150, 2815–2824 (2004).
Cornejo, E., Abreu, N. & Komeili, A. Compartmentalization and organelle formation in bacteria. Curr. Opin. Cell Biol. 26, 132–138 (2014).
Vogel, F., Bornhovd, C., Neupert, W. & Reichert, A. S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175, 237–247 (2006).
Watanabe, M. et al. Attachment of phycobilisomes in an antenna-photosystem I supercomplex of cyanobacteria. Proc. Natl Acad. Sci. USA 111, 2512–2517 (2014).
Iwai, M. et al. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464, 1210–1213 (2010).
Peng, L., Shimizu, H. & Shikanai, T. The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283, 34873–34879 (2008).
Lapuente-Brun, E. et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 (2013).
Katoh, H., Grossman, A. R., Hagino, N. & Ogawa, T. A gene of Synechocystis sp strain PCC 6803 encoding a novel iron transporter. J. Bacteriol. 182, 6523–6524 (2000).
Wang, Q., Hall, C. L., Al-Adami, M. Z. & He, Q. IsiA is required for the formation of photosystem I supercomplexes and for efficient state transition in synechocystis PCC 6803. PLoS ONE 5, e10432 (2010).
Gust, B., Kieser, T. & Chater, K. F. REDIRECT Technology: PCR-targeting System in Streptomyces coelicolor (John Innes Centre, 2002).
Huang, F. et al. Roles of RbcX in carboxysome biosynthesis in the cyanobacterium Synechococcus elongatus PCC7942. Plant Physiol. 179, 184–194 (2019).
Li, M., Semchonok, D. A., Boekema, E. J. & Bruce, B. D. Characterization and evolution of tetrameric photosystem I from the thermophilic cyanobacterium Chroococcidiopsis sp TS-821. Plant Cell 26, 1230–1245 (2014).
Zhang, P. P. et al. Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16, 3326–3340 (2004).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Zhang, P. P. et al. Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24, 1952–1971 (2012).
Faulkner, M. et al. Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes. Nanoscale 9, 10662–10673 (2017).
Acknowledgements
We thank J. Rodriguez-Ramos for support with AFM data analysis and F. Zhao for data analysis. We thank the Liverpool Centre for Cell Imaging and Centre for Proteome Research for technical assistance and provision in AFM imaging, mass spectroscopy and data analysis. We also thank G. F. Dykes and A. Beckett for technical support with EM. This work was supported by the Royal Society University Research Fellowship (nos. UF120411 and URF\R\180030 to L.-N.L.), Royal Society grants (nos. RGF\EA\181061, RGF\EA\180233 and IEC\NSFC\191600 to L.-N.L.), Biotechnology and Biological Sciences Research Council grants (nos. BB/R003890/1, BB/M024202/1, BB/M012441/1 to L.-N.L.), the Queen Mary Principal’s research studentship (to S.W.), the National Science Foundation of China (nos. 31630012, U1706207 and 91851205 to Y.-Z.Z.), the National Key R&D Program of China (no. 2018YFC1406700 to Y.-Z.Z.), the Major Scientific and Technological Innovation Project of Shandong Province (no. 2019JZZY010817 to Y.-Z.Z.), the AoShan Talents Cultivation Program supported by the Pilot National Laboratory for Marine Science and Technology (Qingdao), China (no. 2017ASTCP-OS14 to Y.-Z.Z.), the Taishan Scholars Program of Shandong Province, China (no. tspd20181203 to Y.-Z.Z.), the National Natural Science Foundation of China (nos. 31770128 and 91851103 to Q.W.) and the China Postdoctoral Science Foundation Funded Project (no. 2019M662335 to L.-S.Z.).
Author information
Authors and Affiliations
Contributions
L.-S.Z., Y.-Z.Z. and L.-N.L. conceived the project. L.-S.Z., T.H., S.W., D.M.S., C.W.M. and L.-N.L. performed the research. L.-S.Z., T.H. S.W., D.M.S., Q.W., A.V.R., C.W.M., Y.-Z.Z. and L.-N.L. analysed the data. L.-S.Z., T.H., C.W.M., Y.-Z.Z. and L.-N.L. wrote the manuscript. All authors discussed and commented on the results and the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Figs. 1–18 and references.
Supplementary Data File 1
Statistical source data for Supplementary Fig. 1.
Supplementary Data File 2
Statistical source data for Supplementary Fig. 2.
Supplementary Data File 3
Unprocessed immunoblots and gels corresponding to the immunoblots and gels presented in Supplementary Fig. 2.
Supplementary Data File 4
Statistical source data for Supplementary Fig. 4.
Supplementary Data File 5
Statistical source data for Supplementary Fig. 9.
Supplementary Data File 6
Unprocessed immunoblots and gels corresponding to the immunoblots and gels presented in Supplementary Fig. 9.
Supplementary Data File 7
Statistical source data for Supplementary Fig. 12.
Supplementary Data File 8
Statistical source data for Supplementary Fig. 14.
Supplementary Data File 9
Statistical source data for Supplementary Table 1.
Supplementary Data File 10
Statistical source data for Supplementary Table 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Zhao, LS., Huokko, T., Wilson, S. et al. Structural variability, coordination and adaptation of a native photosynthetic machinery. Nat. Plants 6, 869–882 (2020). https://doi.org/10.1038/s41477-020-0694-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0694-3
This article is cited by
-
Plants and global warming: challenges and strategies for a warming world
Plant Cell Reports (2024)
-
The cytochrome b6f complex: plastoquinol oxidation and regulation of electron transport in chloroplasts
Photosynthesis Research (2024)
-
Energetic robustness to large scale structural fluctuations in a photosynthetic supercomplex
Nature Communications (2023)
-
Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120
Nature Communications (2023)
-
Absolute quantification of cellular levels of photosynthesis-related proteins in Synechocystis sp. PCC 6803
Photosynthesis Research (2023)