Abstract
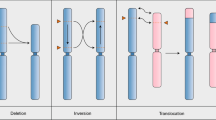
Clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein (Cas) technology has been applied in plant breeding mainly on genes for improving single or multiple traits1,2,3,4. Here we show that this technology can also be used to restructure plant chromosomes. Using the Cas9 nuclease from Staphylococcus aureus5, we were able to induce reciprocal translocations in the Mbp range between heterologous chromosomes in Arabidopsis thaliana. Of note, translocation frequency was about five times more efficient in the absence of the classical non-homologous end-joining pathway. Using egg-cell-specific expression of the Cas9 nuclease and consecutive bulk screening, we were able to isolate heritable events and establish lines homozygous for the translocation, reaching frequencies up to 2.5% for individual lines. Using molecular and cytological analysis, we confirmed that the chromosome-arm exchanges we obtained between chromosomes 1 and 2 and between chromosomes 1 and 5 of Arabidopsis were conservative and reciprocal. The induction of chromosomal translocations enables mimicking of genome evolution or modification of chromosomes in a directed manner, fixing or breaking genetic linkages between traits on different chromosomes. Controlled restructuring of plant genomes has the potential to transform plant breeding.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings are available within the paper and its Supplementary Information, or are available from the corresponding author upon reasonable request.
Code availability
R code for detailed analysis of NGS data is available upon request from the corresponding author.
References
Zhang, Y., Massel, K., Godwin, I. D. & Gao, C. Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210 (2018).
Zhang, Y., Malzahn, A. A., Sretenovic, S. & Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794 (2019).
Schindele, A., Dorn, A. & Puchta, H. CRISPR/Cas brings plant biology and breeding into the fast lane. Curr. Opin. Biotechnol. 61, 7–14 (2020).
Hua, K. et al. Perspectives on the application of genome-editing technologies in crop breeding. Mol. Plant 12, 1047–1059 (2019).
Steinert, J., Schiml, S., Fauser, F. & Puchta, H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 84, 1295–1305 (2015).
Schmidt, C., Schindele, P. & Puchta, H. From gene editing to genome engineering: restructuring plant chromosomes via CRISPR/Cas. aBIOTECH 36, 17 (2019).
Wolter, F., Schindele, P. & Puchta, H. Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19, 176 (2019).
Puchta, H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14 (2005).
Filler Hayut, S., Melamed Bessudo, C. & Levy, A. A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 8, 15605 (2017).
Salomon, S. & Puchta, H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 17, 6086–6095 (1998).
Schimmel, J., Kool, H., van Schendel, R. & Tijsterman, M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 36, 3634–3649 (2017).
Wang, H. & Xu, X. Microhomology-mediated end joining: new players join the team. Cell Biosci. 7, 6 (2017).
Siebert, R. & Puchta, H. Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14, 1121–1131 (2002).
Schmidt, C., Pacher, M. & Puchta, H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 98, 577–589 (2019).
Pacher, M., Schmidt-Puchta, W. & Puchta, H. Two unlinked double-strand breaks can induce reciprocal exchanges in plant genomes via homologous recombination and nonhomologous end joining. Genetics 175, 21–29 (2007).
Rowley, J. D. Chromosome translocations: dangerous liaisons revisited. Nat. Rev. Cancer 1, 245–250 (2001).
Thompson, S. L. & Compton, D. A. Chromosomes and cancer cells. Chromosome Res. 19, 433–444 (2011).
Bunting, S. F. & Nussenzweig, A. End-joining, translocations and cancer. Nat. Rev. Cancer 13, 443–454 (2013).
Lysak, M. A. et al. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl Acad. Sci. USA 103, 5224–5229 (2006).
Gabur, I., Chawla, H. S., Snowdon, R. J. & Parkin, I. A. P. Connecting genome structural variation with complex traits in crop plants. Theor. Appl. Genet. 132, 733–750 (2019).
Huefner, N. D., Mizuno, Y., Weil, C. F., Korf, I. & Britt, A. B. Breadth by depth: expanding our understanding of the repair of transposon-induced DNA double strand breaks via deep-sequencing. DNA Repair 10, 1023–1033 (2011).
Graham, T. G. W., Carney, S. M., Walter, J. C. & Loparo, J. J. A single XLF dimer bridges DNA ends during nonhomologous end joining. Nat. Struct. Mol. Biol. 25, 877–884 (2018).
Wang, Z.-P. et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015).
Wolter, F., Klemm, J. & Puchta, H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 94, 735–746 (2018).
Durr, J., Papareddy, R., Nakajima, K. & Gutierrez-Marcos, J. Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci. Rep. 8, 4443 (2018).
Miki, D., Zhang, W., Zeng, W., Feng, Z. & Zhu, J.-K. CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 9, 1967 (2018).
Kazda, A. et al. Chromosome end protection by blunt-ended telomeres. Genes Dev. 26, 1703–1713 (2012).
Schiml, S., Fauser, F. & Puchta, H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80, 1139–1150 (2014).
Jia, Q., Bundock, P., Hooykaas, P. J. J. & Pater, Sde Agrobacterium tumefaciens T-DNA integration and gene targeting in Arabidopsis thaliana non-homologous end-joining mutants. J. Bot. 2012, 1–13 (2012).
Alonso, J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Park, J., Lim, K., Kim, J.-S. & Bae, S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288 (2017).
Lysak, M. A., Fransz, P. F., Ali, H. B. & Schubert, I. Chromosome painting in Arabidopsis thaliana. Plant J. 28, 689–697 (2001).
Han, Y., Zhang, T., Thammapichai, P., Weng, Y. & Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200, 771–779 (2015).
Acknowledgements
We thank A. Imhof, K. Siegmund, K. Kumke and C. Gäde for technical assistance with performing experiments, T. Wehrle for assisting with cultivation of the plants in the greenhouse, A. Whitbread for critical reading of the manuscript, T. Zundel for providing an R-based script for bioinformatic analysis of NGS data, A. Neubauer for customizing the script to our needs and M. Lysak for the selection of BACs. This research was funded by the European Research Council (ERC) (Grant number ERC-2016-AdG_741306 CRISBREED) to H.P.
Author information
Authors and Affiliations
Contributions
N.B., C.S., M.P. and H.P. designed research; N.B. conducted the research and A.H. performed FISH analysis; N.B., C.S., A.H. and H.P. analysed data; and N.B., A.H. and H.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Keunsub Lee, Kan Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
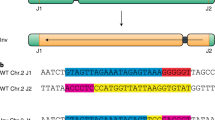
Extended Data Fig. 1 Detailed analysis of NGS data regarding the repair pattern at the junction sites of TL1-2 in T1 generation.
a, Editing efficiency (percentage of modified reads out of total amount of reads) of SaCas9 at the four different target sites. b, Evaluation of NGS data regarding the deletions at the junction sites. Of this, only the reads carrying a deletion were analysed. The lengths of the deletions were divided into three classes (small: 1-10 bp, middle: 11-50 bp, large: > 50 bp). The deletions at the junction sites in Col-0 were mostly of small or middle size, whereas the ku70 mutant showed bigger amounts of large deletions at both junction sites. c, Quantification of the occurrence of microhomologies (MH, ≥ 2 bp) used for junction formation. We separated the reads into three classes: error-free ligated junctions (green), junctions formed without the use of MHs (orange) or with MHs (yellow). In wild type, most junctions were directly ligated without any mutation induction at the junction sites. Of the reads showing mutations at the junction sites, most junctions were joined without the use of MHs, only a minority of junctions were joined using MHs. In contrast, in the ku70 mutant background nearly no error-free ligation occurred and the prevalent repair pattern at the junctions sites showed the use of MHs for joining. d, e, Detailed representation of the 10 most common reads of both junction sites on sequence level in wild type as well as ku70-1.
Extended Data Fig. 2 Molecular and cytological analysis of two independent plant lines (NBE657#24 and NBE669#30) carrying the TL1-2 homozygously.
a, Schematic overview of PCR amplicons (F1-6) of the translocated chromosome parts. A fragment for amplification of around 2.5 kb every 100 kb was designed for each chromosome arm. b, Cropped gel electrophoresis pictures of PCR amplicons of the translocated chromosome parts. Every 100 kb on the translocated chromosome parts, a band could be detected, indicating no information was lost during translocation formation. As PCR controls, genomic DNA of wild type without translocation formation (positive control) and water (NTC = no template control, negative control) were processed at the same time and loaded on different gels for a better overview. c, By phenotypical comparison of 5 week old plants carrying the TL1-2 to the wild type no differences in growth could be documented. Experiments were repeated two times independently with similar results. d, e, Fertility analysis was conducted by measuring the silique length and counting the number of seeds of five biologically independent samples (n = 5). Barplots show the mean values, error bars as mean ± s.d. For both analysed lines carrying the TL1-2, we could not detect any difference to the wild type.
Supplementary information
Supplementary Figure 1
Supplementary Raw Data 1–5 and Tables 1–7.
Source data
Source Data Fig. 3
Microscopy pictures TL1-2, unmerged and merged.
Source Data Fig. 4
Microscopy pictures TL1-5, unmerged and merged.
Source Data Extended Data Fig. 2
Unprocessed gel pictures.
Rights and permissions
About this article
Cite this article
Beying, N., Schmidt, C., Pacher, M. et al. CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 6, 638–645 (2020). https://doi.org/10.1038/s41477-020-0663-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0663-x
This article is cited by
-
Genome editing based trait improvement in crops: current perspective, challenges and opportunities
The Nucleus (2024)
-
Crop bioengineering via gene editing: reshaping the future of agriculture
Plant Cell Reports (2024)
-
Applications of CRISPR/Cas genome editing in economically important fruit crops: recent advances and future directions
Molecular Horticulture (2023)
-
Genome-edited foods
Nature Reviews Bioengineering (2023)
-
Hidden prevalence of deletion-inversion bi-alleles in CRISPR-mediated deletions of tandemly arrayed genes in plants
Nature Communications (2023)