Abstract
Directional intercellular transport of the phytohormone auxin mediated by PIN-FORMED (PIN) efflux carriers has essential roles in both coordinating patterning processes and integrating multiple external cues by rapidly redirecting auxin fluxes. PIN activity is therefore regulated by multiple internal and external cues, for which the underlying molecular mechanisms are not fully elucidated. Here, we demonstrate that 3′-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE1 (PDK1), which is conserved in plants and mammals, functions as a molecular hub that perceives upstream lipid signalling and modulates downstream substrate activity through phosphorylation. Using genetic analysis, we show that the loss-of-function Arabidopsis pdk1.1 pdk1.2 mutant exhibits a plethora of abnormalities in organogenesis and growth due to defective polar auxin transport. Further cellular and biochemical analyses reveal that PDK1 phosphorylates D6 protein kinase, a well-known upstream activator of PIN proteins. We uncover a lipid-dependent phosphorylation cascade that connects membrane-composition-based cellular signalling with plant growth and patterning by regulating morphogenetic auxin fluxes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data for Figs. 1–6, and Extended Figs. 2–4, 7, 9 and 10 are provided with the paper. Sequencing data from this Article is provided in the Arabidopsis Genome Initiative databases under the following accession numbers: PIN1 (AT1G73590), PIN2 (AT5G57090), PIN3 (AT1G70940), PIN4 (AT2G01420), PIN7 (AT1G23080), PDK1.1 (AT5G04510), PDK1.2 (AT3G10540), D6PK (AT5G55910), D6PKL1 (AT4G26610), D6PKL2 (AT5G47750), D6PKL3 (AT3G27580), PID (AT2G34650), WAG1 (AT1G53700) and WAG2 (AT3G14370). All data necessary to evaluate the conclusions in the paper or the Supplementary Information are available from the corresponding authors on request.
Change history
12 June 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41477-020-0719-y
References
Swarup, K. et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954 (2008).
Petrášek, J. et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918 (2006).
Adamowski, M. & Friml, J. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32 (2015).
Geisler, M. et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44, 179–194 (2005).
Armengot, L., Marquès-Bueno, M. M. & Jaillais, Y. Regulation of polar auxin transport by protein and lipid kinases. J. Exp. Bot. 67, 4015–4037 (2016).
Friml, J. et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306, 862–865 (2004).
Dhonukshe, P. et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137, 3245–3255 (2010).
Grones, P. et al. PID/WAG-mediated phosphorylation of the Arabidopsis PIN3 auxin transporter mediates polarity switches during gravitropism. Sci. Rep. 8, 10279 (2018).
Zourelidou, M. et al. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136, 627–636 (2009).
Zourelidou, M. et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3, e02860 (2014).
Marhava, P. et al. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558, 297–300 (2018).
Jia, W. et al. Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biol. 14, e1002550 (2016).
Dory, M. et al. Coevolving MAPK and PID phosphosites indicate an ancient environmental control of PIN auxin transporters in land plants. FEBS Lett. 592, 89–102 (2018).
Rigó, G. et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 25, 1592–1608 (2013).
Michniewicz, M. et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056 (2007).
Dai, M. et al. A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24, 2497–2514 (2012).
Guo, X. et al. TYPE-ONE PROTEIN PHOSPHATASE4 regulates pavement cell interdigitation by modulating PIN-FORMED1 polarity and trafficking in Arabidopsis. Plant Physiol. 167, 1058–1075 (2015).
Weller, B. et al. Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc. Natl Acad. Sci. USA 114, E887–E896 (2017).
Laetitia, M. et al. A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat. Plants 2, 16089 (2016).
Wang, P. et al. Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED 2 auxin efflux transporter in response to salt stress. Plant Cell 31, 250–271 (2019).
Barbosa, I. C. R. et al. Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143, 4687–4700 (2016).
Pearce, L. R., Komander, D. & Alessi, D. R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 (2010).
Rintelen, F., Stocker, H., Thomas, G. & Hafen, E. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15020–15025 (2001).
Lawlor, M. A. et al. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 21, 3728–3738 (2002).
Deak, M., Casamayor, A., Currie, R. A., Peter Downes, C. & Alessi, D. R. Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett. 451, 220–226 (1999).
Rentel, M. C. et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427, 858–861 (2004).
Anthony, R. G., Khan, S., Costa, J., Pais, M. S. & Bögre, L. The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J. Biol. Chem. 281, 37536–37546 (2006).
Camehl, I. et al. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 7, e1002051 (2011).
Anthony, R. G. et al. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581 (2004).
Rademacher, E. H. & Offringa, R. Evolutionary adaptations of plant AGC kinases: from light signaling to cell polarity regulation. Front. Plant Sci. 3, 250 (2012).
Zegzouti, H. et al. Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J. Biol. Chem. 281, 35520–35530 (2006).
Scholz, S. et al. The AGC protein kinase UNICORN controls planar growth by attenuating PDK1 in Arabidopsis thaliana. PLoS Genet. 15, e1007927 (2019).
Xiao, Y. & Offringa, R. PDK1 regulates auxin transport and Arabidopsis vascular development through AGC1 kinase PAX. Nat. Plants https://doi.org/10.1038/s41477-020-0650-2 (2020).
Friml, J. et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673 (2002).
Band, L. R. et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl Acad. Sci. USA 109, 4668–4673 (2012).
Sabatini, S. et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472 (1999).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153 (2003).
Prát, T. et al. WRKY23 is a component of the transcriptional network mediating auxin feedback on PIN polarity. PLoS Genet. 14, e1007177 (2018).
Zadnikova, P. et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137, 607–617 (2010).
Stepanova, A. N. et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008).
Friml, J., Wiśniewska, J., Benková, E., Mendgen, K. & Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002).
Gray, W. M., Ostin, A., Sandberg, G., Romano, C. P. & Estelle, M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl Acad. Sci. USA 95, 7197–7202 (2002).
Franklin, K. A. et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl Acad. Sci. USA 108, 20231–20235 (2011).
Parry, G. & Estelle, M. Auxin receptors: a new role for F-box proteins. Curr. Opin. Cell Biol. 18, 152–156 (2006).
Brumos, J. et al. Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47, 306–318 (2018).
Luschnig, C., Gaxiola, R. A., Grisafi, P. & Fink, G. R. EIR1, a root specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187 (1998).
Swarup, R. et al. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7, 1057–1065 (2005).
Zegzouti, H., Anthony, R. G., Jahchan, N., Bogre, L. & Christensen, S. K. Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl Acad. Sci. USA 103, 6404–6409 (2006).
Zhang, J., Nodzynski, T., Pencik, A., Rolcik, J. & Friml, J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl Acad. Sci. USA 107, 918–922 (2010).
Van Leeuwen, W., Ökrész, L., Bögre, L. & Munnik, T. Learning the lipid language of plant signalling. Trends Plant Sci. 9, 378–384 (2004).
Geldner, N. et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230 (2003).
Noack, L. C. & Jaillais, Y. Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Curr. Opin. Plant Biol. 40, 22–33 (2017).
Platre, M. P. et al. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364, 57–62 (2019).
Mei, Y., Jia, W., Chu, Y. & Xue, H. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res. 22, 581–597 (2011).
Stenzel, I. et al. Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. Plant Cell 25, 4894–4911 (2013).
Tejos, R. et al. Bipolar plasma membrane distribution of phosphoinositides and their requirement for auxin-mediated cell polarity and patterning in Arabidopsis. Plant Cell 26, 2114–2128 (2014).
Gao, H. B., Chu, Y. J. & Xue, H. W. Phosphatidic acid (PA) binds PP2AA1 to regulate PP2A activity and PIN1 polar localization. Mol. Plant 6, 1692–1702 (2013).
Zhang, J. et al. Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev. Cell 20, 855–866 (2011).
Xu, J. et al. A molecular framework for plant regeneration. Science 311, 385–388 (2006).
Benková, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003).
Xu, J. & Scheres, B. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17, 525–536 (2005).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Liu, W., Xu, Z. H., Luo, D. & Xue, H. W. Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36, 189–202 (2003).
Rook, F. et al. Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 15, 253–263 (1998).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Baster, P. et al. SCFTIR1/AFB-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 32, 260–274 (2013).
Abas, L. et al. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8, 249–256 (2006).
Lewis, D. R. & Muday, G. K. Measurement of auxin transport in Arabidopsis thaliana. Nat. Protoc. 4, 437–451 (2009).
Tan, S. et al. Salicylic acid targets protein phosphatase 2A to attenuate growth in plants. Curr. Biol. 30, 381–395 (2020).
Acknowledgements
We thank C. Schwechheimer and B. Scheres for sharing published materials; M. Glanc for providing pET28a-PIN2/3 plasmids; X. Gao for help with SEM imaging, L. Rodriguez for advice on co-IP; staff at the bioimaging and life science facilities of IST Austria for continuous service and assistance; and the Nottingham Arabidopsis Stock Centre (NASC) and the Arabidopsis Biological Resource Centre (ABRC) for providing T-DNA insertional mutants. J.P. acknowledges the support from imaging facility of IEB CAS. The research leading to these results has received funding from Chinese Ten-Thousand Talent Program (to H.-W.X.) and the European Union’s Horizon2020 program (ERC grant agreement no. 742985, to J.F.). S.T. was funded by a European Molecular Biology Organization (EMBO) long-term postdoctoral fellowship (ALTF 723–2015). X.Z. was supported by a PhD scholarship from China Scholarship Council.
Author information
Authors and Affiliations
Contributions
S.T., J.F. and H.-W.X. designed experiments. S.T., X.Z., W.K. and X.-L.Y. performed experiments. G.M. provided [32P]ATP and helped with kinase assays. J.P., Z.V. and R.F. performed experiments in BY-2 cells. S.T., J.F. and H.-W.X. analysed and interpreted the data. S.T., J.F. and H.-W.X. wrote the manuscript with input from other co-authors, and all of the authors read and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Expression pattern of PDK1.1 and PDK1.2.
GUS staining of pPDK1.1::GUS and pPDK1.2::GUS lines indicated that PDK1.1 and PDK1.2 were expressed in the vascular tissues in both roots and shoots at various developmental stages, including young seedlings (a, f; 7 days), root stele (b, g; 7 days), columella cells (c, h, only with expression detected for PDK1.1; 7 days), lateral root primordia (d, i; 12 days), and dark-grown seedlings (e, j; 4 days). Representative images of three independent homozygous lines were shown. Scale bars, 1 mm.
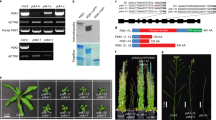
Extended Data Fig. 2 Identification of Arabidopsis pdk1.1 and pdk1.2 T-DNA insertional mutants.
a, Schematic representation of PDK1.1 and PDK1.2 genes and positions of T-DNA insertions for pdk1.1 and pdk1.2. Introns, exons, and non-coding regions are indicated by lines, black, or blank boxes respectively. Positions of primers are indicated. b, Identification of homozygous pdk1.1 and pdk1.2 mutants. Genomic DNA of pdk1.1 and pdk1.2 mutants was used as templates for PCR amplification. Homozygous lines have a single amplified DNA fragment when using LBa1/pdk1.1-RP or LBa1/pdk1.2-RP primers. n = 5 biologically independent experiments, with similar results obtained. c, qRT-PCR analysis confirmed the deficient expression of PDK1.1 and PDK1.2 genes in pdk1.1 and pdk1.2 mutants, respectively. Total RNA of 7-day-old WT, pdk1.1 and pdk1.2 seedlings was extracted, reversely transcribed, and then used for analysis. ACTIN7 was amplified and used as an internal reference to normalize the expression of PDK1.1 and PDK1.2, and the mean value in Col-0 was set as “1”. The experiments were biologically repeated for 3 times. Dots represent individual samples, and lines indicate mean ± s.d.. P values were calculated by a Welch’s two-tailed t-test.
Extended Data Fig. 3 Deficiency of PDK1.1 and PDK1.2 impaired the hypocotyl gravitropism under dark and phototropism towards directional light.
a, Deficiency of PDK1.1 and PDK1.2 promoted the radial growth in the root columella cell region. Transverse view of the root columella cells by CLSM. Left, a schematic image to show the positon for the transverse view; middle, Col-0; right, pdk1.1 pdk1.2. Scale bars, 20 µm. n = 3 biologically independent experiments, with similar results obtained. b, Deficiency of PDK1.1 and PDK1.2 gave rise to more columns, but not layers of, root columella cells. Quantification is based on CLSM images of PI-stained roots. Layer numbers were counted for both undifferentiated and differentiated columella cells. Dots represent individual plants, and lines indicate mean ± s.d.. n = 18, 26, 18, and 26, from left to right, respectively. P values were calculated by a Welch’s two-tailed t-test. c–e, Deficiency of PDK1.1 and PDK1.2 impaired root and shoot gravitropic response in the dark. Etiolated seedlings of Col-0, pdk1.1, pdk1.2, and pdk1.1 pdk1.2 were grown under dark for 90 h and a representative photo was shown (c). Scale bar, 5 mm. n = 5 biologically independent experiments, with similar results obtained (c). (d), Root tip angles are shown as polar bar charts. n = 61, 62, 69, and 71, respectively. (e), Hypocotyl angles are shown as polar bar charts. n = 51, 50, 59, and 68, respectively. f–g, pdk1.1 pdk1.2 showed defects in phototropism. Seedlings of Col-0, pdk1.1, pdk1.2, and pdk1.1 pdk1.2 were grown under dark for 90 h, exposed to white light for 24 h, and were then subjected to directional white light in a box covered with aluminium foil from the other sides. A representative photo is shown (f). Scale bar, 5 mm. n = 3 biologically independent experiments, with similar results obtained (f). (g), hypocotyl angles are shown by polar bar charts. n = 56, 62, 67, and 62, respectively. P values were calculated by a Welch’s two-tailed t-test, and also by a further F-test to indicate differences of variances (d, e, and g).
Extended Data Fig. 4 Deficiency of PDK1.1 and PDK1.2 impaired the normal development of the apical hook and high temperature-induced hypocotyl elongation.
a, b, Observation (a, scale bar, 5 mm) and quantification (b) showed that etiolated seedlings (90 h) of pdk1.1 pdk1.2 exhibited less tight apical hooks. n = 5 biologically independent experiments, with similar results obtained (a). n = 14, 18, 12 and 17 seedlings for Col-0, pdk1.1, pdk1.2, and pdk1.1 pdk1.2 respectively (b). c, Etiolated seedlings of pdk1.1 pdk1.2 exhibited comparably long hypocotyls to Col-0. Seedlings were grown under dark for 90 h. n = 20, 21, 20 and 17 seedlings for Col-0, pdk1.1, pdk1.2, and pdk1.1 pdk1.2 respectively. d, Etiolated seedlings of pdk1.1 pdk1.2 did not form exaggerated apical hooks in the presence of ACC. Seedlings of Col-0, pdk1.1, pdk1.2, and pdk1.1 pdk1.2 were grown under dark for 90 h in the absence or presence of ACC (10 µM). Scale bars, 500 µm. n = 3 biologically independent experiments, with similar results obtained. e–f, pdk1.1 pdk1.2 showed defects in high temperature-induced hypocotyl elongation. Seedlings of Col-0, pdk1.1, pdk1.2 and pdk1.1 pdk1.2 were grown under light at 22 °C (n = 16, 20, 18 and 22 respectively) or 29 °C (n = 20, 21, 17 and 18) for 5 days, and hypocotyl elongation was observed (e, scale bar, 1 cm) and quantified (e). Hypocotyl length was measured by the Image J program and shown as mean ± s.d. (left) or relative length by setting the hypocotyl length of Col-0 and pdk1.1 pdk1.2 at 22 °C as “1”, respectively (right). Dots represent individual plants, and lines indicate mean ± s.d.. Different letters represent significant difference, P < 0.05, by one-way ANOVA with a Tukey multiple comparison test, and P values are shown for each genotype compared with Col-0 (b, c, and f).
Extended Data Fig. 5 Loss of function of PDK1.1 and PDK1.2 impaired auxin distribution.
Observation of the auxin responsive reporter DR5rev::GFP by CLSM indicated a dramatic decrease of the auxin maxima in pdk1.1 pdk1.2 (e–h) compared with Col-0 (a–d). Fused cotyledon exhibiting two sites of auxin maxima in light-grown 7-day-old seedlings of pdk1.1 pdk1.2 (e) compared with one of Col-0 (a); roots of light-grown 10-day-old seedlings (b and f); apical hooks of 10 µM ACC treated 4-day-old etiolated seedlings (c and g); roots of 10 µM ACC treated 4-day-old etiolated seedlings (d and h). Scale bars, 200 µm. n = 3 biologically independent experiments, with similar results obtained.
Extended Data Fig. 6 Deficiency of PDK1.1 and PDK1.2 did not affect the polarity of PIN proteins.
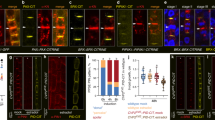
a, Deficiency of PDK1.1 and PDK1.2 did not change the polarity of PIN1-YFP. Four-day-old seedlings of pPIN1::PIN1-YFP in Col-0 and pPIN1::PIN1-YFP in pdk1.1 pdk1.2 were imaged by CLSM. Scale bars, 20 µm. n = 2 biologically independent experiments, with similar results obtained (a-g). b, Deficiency of PDK1.1 and PDK1.2 did not change the polarity of PIN2-GFP. Four-day-old seedlings of pPIN2::PIN2-GFP in Col-0 and pPIN2::PIN2-GFP in pdk1.1 pdk1.2 were imaged by CLSM. Scale bars, 20 µm. c–f, Deficiency of PDK1.1 and PDK1.2 did not change the polarity of PIN3-GFP. Four-day-old seedlings of pPIN3::PIN3-GFP in Col-0 and pPIN3::PIN3-GFP in pdk1.1 pdk1.2 were imaged by CLSM. (c) subcellular localisation of PIN3-GFP; (d) an amplified view of PIN3-GFP in the root stele; (e) a close view of PIN3-GFP in root columella cells; (f) a 3D-projection of PIN3-GFP localisation in root columella cells. The “Green Fire Blue” LUT was used for photo visualization based on fluorescence intensity by Fiji. Scale bars, 20 µm. g, A transverse view of the root columella cells revealed the overproliferation, with an enlarged region expressing PIN3-GFP. Four-day-old seedlings of pPIN3::PIN3-GFP in Col-0 and pPIN3::PIN3-GFP in pdk1.1 pdk1.2 were stained with PI, and imaged by CLSM, at the position as marked in the left image. Scale bars, 20 µm.
Extended Data Fig. 7 Analysis of PDK1 transgenic lines.
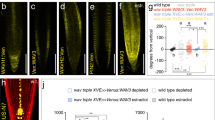
a, Expression of PDK1.1 or PDK1.2 (p35S::Venus-PDK1.1, p35S::Venus-PDK1.2, p35S::mCherry-PDK1.1 and p35S::mCherry-PDK1.2) rescued the growth defects of pdk1.1 pdk1.2. Adult plants (25-day-old) were observed and representative photos were shown. Scale bar, 2 cm. b, Western blot analysis verified the PDK1.1 or PDK1.2 expression (p35S::Venus-PDK1.1 and p35S::Venus-PDK1.2) in pdk1.1 pdk1.2, respectively. Seven-day-old T3 homozygous seedlings were used for protein extraction and subjected to analysis. Upper panel, anti-GFP (1:2000); lower panel, Ponceau staining. c, Western blot verified the PDK1.1 or PDK1.2 expression (p35S::mCherry-PDK1.1 and p35S::mCherry-PDK1.2) in pdk1.1 pdk1.2, respectively. Seven-day-old T3 homozygous seedlings were used for protein extraction and subjected to analysis. Upper panel, anti-RFP (1:2000); lower panel, Ponceau staining. d–e, mCherry-fused PDK1.1 (d) and PDK1.2 (e) localised to both cytoplasm and the basal side of PM. Four-day-old seedlings of p35S::mCherry-PDK1.1 and p35S::mCherry-PDK1.2 were imaged by CLSM. Open arrowheads indicated the basal polar localisation. Scale bar, 10 µm. f–i, Subcellular localisation of Venus-fused PDK1.1N (f, cytoplasm), Venus-fused PDK1.1C (g, both cytoplasm and nucleus), Venus-fused PDK1.2N (h, cytoplasm), and Venus-fused PDK1.2C (i, both cytoplasm and nucleus). Four-day-old seedlings expressing corresponding fusion proteins were imaged by CLSM. Scale bar, 10 µm. n = 4, 2, 3, and 3 biologically independent experiments for (a), (b), (c), and (d) respectively, with similar results obtained.
Extended Data Fig. 8 Functional pPDK1.1::Venus-PDK1.1 localised at both cytoplasm and the basal side of PM.
a, pPDK1.1::Venus-PDK1.1 rescued the growth defects of pdk1.1 pdk1.2. 25-day-old adult plants were observed and representative photos are shown. Scale bar, 2 cm. b, pPDK1.1::Venus-PDK1.1 rescued the lateral root defects of pdk1.1 pdk1.2. 10-day-old seedlings were observed and representative photos are shown. Scale bar, 2 cm. c, pPDK1.1::Venus-PDK1.1 rescued the defects of pdk1.1 pdk1.2 in the hypocotyl gravitropic response. 4-day-old etiolated seedlings were observed and representative photos are shown. Scale bar, 2 cm. d, Venus-fused PDK1.1 localised to both cytoplasm and PM, especially with a predominant presence at the basal side of PM. Four-day-old seedlings of pPDK1.1::Venus-PDK1.1 were imaged by CLSM. Open arrowheads indicate the basal polar localisation. The “Green Fire Blue” LUT was used for photo visualization based on fluorescence intensity by Fiji. Scale bar, 20 µm. e, Venus-PDK1.1 localized to cytoplasm (upper image) in interphase tobacco BY-2 cells, and the association with PM was obvious only during cytokinesis (arrow in lower image). An XVE>>Venus-PDK1.1 line was induced for 48 h with 1 μM β-estradiol and were then imaged by spinning disk confocal microscope. Scale bar, 10 μm. n = 3, 3, 2, 3, and 2 biologically independent experiments for (a), (b), (c), (d), and (e) respectively, with similar results obtained.
Extended Data Fig. 9 mCherry-D6PKs co-localised with Venus-PDK1.1 at the basal side of PM.
a, mCherry-fused D6PKs localised to the basal side of PM. Four-day-old seedlings of p35S::mCherry-D6PK/D6PKLs (short as D0 to D3) were imaged by CLSM. The “mpl-inferno” LUT was used for photo visualization based on fluorescence intensity by Fiji. Scale bars, 10 µm. b, Western blot verified the mCherry-D0~D3 protein level (35S::mCherry-D0~D3) in Col-0, respectively. Seven-day-old T3 homozygous seedlings were used for protein extraction and subjected to Western blot analysis. Upper panel, anti-RFP (1:1000), short exposure (0.5 sec); medium panel, anti-RFP (1:1000), long exposure (5 sec, for low expression of mCherry-D3); lower panel, Ponceau staining. c, Venus-PDK1.1 co-localised with mCherry-D6PKL2 and mCherry-D6PKL3 at the basal side of PM. Four-day-old seedlings of p35S::mCherry-D6PKL2/ p35S::Venus-PDK1.1 and p35S::mCherry-D6PKL3/ p35S::Venus-PDK1.1 were imaged by CLSM. Scale bar, 10 µm. d, In vitro kinase assay with [32P]-ATP revealed that GST-PDK1.2-conducted phosphorylation of D6PK facilitates its activity towards PIN-HL phosphorylation. Upper panel, autoradiography; lower panel, CBB staining. e, In vitro kinase assay with [32P]-ATP revealed that GST-PDK1.2-conducted full phosphorylation and activation of D6PK, towards His-PIN1-HL phosphorylation, required the phosphorylation at S345 for D6PK. Upper panel, autoradiography of 32P; lower panel, CBB staining. n = 3, 2, 2, 3, and 3 biologically independent experiments for (a), (b), (c), (d), and (e) respectively, with similar results obtained.
Extended Data Fig. 10 Overexpression of Venus-D6PKS345D rescued the defects of lateral root formation and hypocotyl gravitropism in pdk1.1 pdk1.2.
a, b, 35S::Venus-D6PKS345D (L3 as a representative line) rescued the lateral root defects of pdk1.1 pdk1.2. Nine-day-old seedlings were observed. n = 3 biologically independent experiments, with similar results obtained. A representative photo is shown in (a, scale bar, 2 cm), and the lateral root number was quantified (b). Dots represent individual plants, and lines indicate mean ± s.d.. n = 15, 15, 14 and 14 individual seedlings for Col-0, pdk1.1 pdk1.2, 35S::Venus-D6PKS345D in pdk1.1 pdk1.2 (L3), and 35S::Venus-D6PKS345D in pdk1.1 pdk1.2 (L5), respectively. Different letters represent significant difference, P < 0.05, by one-way ANOVA with a Tukey multiple comparison test, and P values are shown for each genotype compared with Col-0. c, The subcellular localisations of Venus-D6PK, Venus-D6PKS345A, and Venus-D6PKS345D exhibit difference in PM targeting, but not show difference in the basal polarity. Four-day-old seedlings of p35S::Venus-D6PK, p35S::Venus-D6PKS345A, and p35S::Venus-D6PKS345D, in Col-0 and pdk1.1 pdk1.2 respectively, were imaged by CLSM. The “Green Fire Blue” LUT was used for photo visualization based on fluorescence intensity by Fiji. Representative photos of at least three independent transgenic lines were shown. Scale bars, 10 µm. n = 2 biologically independent experiments, with similar results obtained.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Tables 1–7 and references for the Supplementary Information.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Tan, S., Zhang, X., Kong, W. et al. The lipid code-dependent phosphoswitch PDK1–D6PK activates PIN-mediated auxin efflux in Arabidopsis. Nat. Plants 6, 556–569 (2020). https://doi.org/10.1038/s41477-020-0648-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0648-9