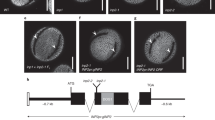
Abstract
The aperture on the pollen surface provides an exit for the emerging pollen tube. Apertures exhibit huge morphological variation across plant species—grasses, including rice, possess a complex aperture consisting of an annulus and an operculum—but little is known about how this species-specific cell-surface pattern forms. Here, we report a lectin receptor-like kinase in Oryza sativa, OsDAF1, which is essential for annulus formation and thus for fertility. OsDAF1 is evenly distributed in early microsporocytes but localizes to the distal pre-aperture site at the tetrad stage. We further reveal that the rice orthologue of a key aperture factor in Arabidopsis, OsINP1, has conserved and diversified roles in rice aperture formation. Disruption of OsINP1 prevents formation of the aperture, precluding pollen-tube germination. Furthermore, our results demonstrate that OsINP1 is required for polarization of OsDAF1 via direct protein interaction, suggesting that OsINP1 has an additional role in the formation of annulus that is absent in Arabidopsis. Our study reveals the importance of the aperture for rice grain yield and reveals mechanisms controlling pollen aperture development in cereal species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source Data for Fig. 5 and Extended Data Fig. 10 are provided with the paper. Sequence data from this article can be found in the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu) under accession numbers LOC_Os02g26160 (OsDAF1) and LOC_Os02g44250 (OsINP1). The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Shi, J., Cui, M., Yang, L., Kim, Y. J. & Zhang, D. Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20, 741–753 (2015).
Ariizumi, T. & Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62, 437–460 (2011).
Li, F.-S., Phyo, P., Jacobowitz, J., Hong, M. & Weng, J.-K. The molecular structure of plant sporopollenin. Nat. Plants 5, 41–46 (2019).
Quilichini, T. D., Grienenberger, E. & Douglas, C. J. The biosynthesis, composition and assembly of the outer pollen wall: a tough case to crack. Phytochemistry 113, 170–182 (2015).
Furness, C. A. & Rudall, P. J. Pollen aperture evolution—a crucial factor for eudicot success? Trends Plant Sci. 9, 154–158 (2004).
Edlund, A. F. Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16, S84–S97 (2004).
McCormick, S. Control of male gametophyte development. Plant Cell 16, 142–154 (2004).
Zhang, D., Luo, X. & Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38, 379–390 (2011).
Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434 (2005).
Wang, R. & Dobritsa, A. Exine and aperture patterns on the pollen surface: their formation and roles in plant reproduction. Annu. Plant Rev. 1, 1–40 (2018).
Dobritsa, A. A. & Reeder, S. H. Formation of pollen apertures in Arabidopsis requires an interplay between male meiosis, development of INP1-decorated plasma membrane domains, and the callose wall. Plant Signal. Behav. 12, e1393136 (2017).
Reeder, S. H., Lee, B. H., Fox, R. & Dobritsa, A. A. A ploidy-sensitive mechanism regulates aperture formation on the Arabidopsis pollen surface and guides localization of the aperture factor INP1. PLoS Genet. 12, e1006060 (2016).
Albert, B., Ressayre, A. & Nadot, S. Correlation between pollen aperture pattern and callose deposition in late tetrad stage in three species producing atypical pollen grains. Am. J. Bot. 98, 189–196 (2011).
Nadot, S., Forchioni, A., Penet, L., Sannier, J. & Ressayre, A. Links between early pollen development and aperture pattern in monocots. Protoplasma 228, 55–64 (2006).
Toghranegar, Z., Nadot, S. & Albert, B. Variation of microsporogenesis in monocots producing monosulcate pollen grains. Ann. Bot. 112, 135–139 (2013).
Albert, B. et al. Effect of aperture number on pollen germination, survival and reproductive success in Arabidopsis thaliana. Ann. Bot. 121, 733–740 (2018).
Prieu, C. et al. Aperture number influences pollen survival in Arabidopsis mutants. Am. J. Bot. 103, 452–459 (2016).
Köhler, E. & Lange, E. A contribution to distinguishing cereal from wild grass pollen grains by LM and SEM. Grana 18, 133–140 (1979).
El-Ghazaly, G. & Jensen, W. A. Studies of the development of wheat (Triticum aestivum) pollen. Grana 25, 1–29 (1986).
Dobritsa, A. A. & Coerper, D. The novel plant protein INAPERTURATE POLLEN1 marks distinct cellular domains and controls formation of apertures in the Arabidopsis pollen exine. Plant Cell 24, 4452–4464 (2012).
Li, P. et al. INP1 involvement in pollen aperture formation is evolutionarily conserved and may require species-specific partners. J. Exp. Bot. 69, 983–996 (2018).
Dobritsa, A. A., Kirkpatrick, A. B., Reeder, S. H., Li, P. & Owen, H. A. Pollen aperture factor INP1 acts late in aperture formation by excluding specific membrane domains from exine deposition. Plant Physiol. 176, 326–339 (2018).
Lee, B. H. et al. Arabidopsis protein kinase D6PKL3 is involved in the formation of distinct plasma membrane aperture domains. Plant Cell 30, 2038–2056 (2018).
Chen, L. et al. Isolation and genetic analysis for rice mutants treated with 60Co γ-ray. J. Xiamen Univ. 45, 82–85 (2006).
Zhang, D. B. & Wilson, Z. A. Stamen specification and anther development in rice. Chin. Sci. Bull. 54, 2342–2353 (2009).
Vaid, N., Macovei, A. & Tuteja, N. Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol. Plant. 6, 1405–1418 (2013).
Chen, C. et al. The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24, 1256–1270 (2012).
Labbé, J. et al. Mediation of plant–mycorrhizal interaction by a lectin receptor-like kinase. Nat. Plants 5, 676–680 (2019).
Wan, J. et al. A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol. Biol. 67, 469–482 (2008).
Shao, W. & Dong, J. Polarity in plant asymmetric cell division: Division orientation and cell fate differentiation. Dev. Biol. 419, 121–131 (2016).
Zhang, Y., Guo, X. & Dong, J. Phosphorylation of the polarity protein BASL differentiates asymmetric cell fate through MAPKs and SPCH. Curr. Biol. 176, 139–148 (2017).
Bellande, K., Bono, J. J., Savelli, B., Jamet, E. & Canut, H. Plant lectins and lectin receptor-like kinases: how do they sense the outside? Int. J. Mol. Sci. 18, 1164 (2017).
Peng, X. et al. Lectin receptor kinase OsLecRK‐S.7 is required for pollen development and male fertility. J. Int. Plant Biol. https://doi.org/10.1111/jipb.12897 (2020).
Liu, H. et al. Genetic analysis and mapping of rice (Oryza sativa L.) male-sterile (OsMS-L) mutant. Chin. Sci. Bull. 50, 122–125 (2005).
Li, N. et al. The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014 (2006).
Liu, L. et al. Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLoS Genet. 12, 1–23 (2016).
Wang, C. et al. Resolvase OsGEN1 mediates DNA repair by homologous recombination. Plant Physiol. 173, 1316–1329 (2017).
Zhang, H. et al. Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22, 672–689 (2010).
Hu, B. et al. Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413 (2019).
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100902), the National Transgenic Major Program Grant (2016ZX08009-003), National Natural Science Foundation of China (31670309), the Innovative Research Team, Ministry of Education, and 111 Project (Grant B14016).
Author information
Authors and Affiliations
Contributions
W.L. and D.Z. designed the project. X.Z. and G.Z. performed most of the experiments. H.Y. performed the map-based cloning. Q.T. performed the rice transformation. L.Z. and Q.T. performed the TEM sections. W.L., X.Z. and N.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Anna Dobritsa, Dazhong Zhao and the other, anonymous, reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Phenotypes and complementation of osdaf1 mutants.
a–c, Comparisons of wild type (WT) and osdaf1-1 mature plants (a), flowering panicles (b) and spikelets (c). le, lemma; pa; palea; gl, glume. d, e, Comparisons of WT, osdaf1-1, osdaf1-2, osdaf1-3 and gOsDAF1 osdaf1-1 flowers (d) and mature panicles (e). f, The proportion of viable pollen produced by wild-type and mutant plants displayed as box plots, showing the first and third quartiles, split by the median and extended to minimum and maximum values. For each line, 50 independent replicates, each of at least 100 pollen grains, were counted. The phenotypes in a–e were observed at least three times independently with similar results. Scale bars, 10 cm (a), 1 cm (b, e) and 1 mm (c, d).
Extended Data Fig. 2 Anther transverse sections of wild-type and osdaf1 plants.
Transverse sections of wild-type, osdaf1-1 and osdaf1-2 anthers at different developmental stages. Phenotypes were observed three times independently with similar results. Scale bars, 80 µm.
Extended Data Fig. 3 Pollen germination assay of wild-type and osdaf1 plants.
In vivo (a) and in vitro (b) pollen germination assays of wild-type and osdaf1-1 pollen grains. Arrowheads in a indicate the ovule; arrows in b indicate mutant pollen grains that failed to germinate. These experiments were repeated three times independently with similar results. Scale bars, 200 µm (a) and 50 µm (b).
Extended Data Fig. 4 TEM observation of wild-type and osdaf1-1 pollen aperture areas.
Microspores (left panel) and enlarged aperture areas (right panel) from wild-type (upper panel) and osdaf1-1 (lower panel) at stage 8b (a), stage 9 (b), stage 10a (c), stage 10b (d), stage 11 (e), and stage 12 (f). A, annulus; N, nexine; S, sexine; O, operculum; PM, plasma membrane; MSP, microspore; MP, mature pollen; F, fibrillar-granular layer; Z, Zwischenkörper layer; In, intine. Arrows indicate the trilamellated structures in annulus. Arrowheads indicate the missing annulus structure in osdaf1-1 pollen. These phenotypes were observed three times independently with similar results. Scale bars are indicated.
Extended Data Fig. 5 Identification of OsDAF1.
a, Fine mapping of OsDAF1. Positions and numbers of recombinants of each molecular marker are indicated. The OsDAF1 locus is mapped to a 4138 kb region between Y3’ and YH42-5. AP005534 and AP004850 are accession numbers of BAC clones containing the two molecular markers. b, qRT-PCR analysis of OsDAF1. Quantification was normalized to the expression of the internal control ACTIN. n = 3 biologically independent samples of 50 µg anther from each stage. Data are mean ± SD; dots show data distribution.
Extended Data Fig. 6 Subcellular localizations of OsDAF1.
Subcellular localizations of OsDAF1-eYFP protein in tobacco leaf epidermal cells (left panel) and rice protoplasts (right panel). The subcellular localizations were observed three times independently with similar results. Scale bars are indicated.
Extended Data Fig. 7 Phenotypes of osinp1 mutants.
a, Comparisons of WT, osinp1-1, osinp1-2 and osinp1 osdaf1 flowers (a) and mature panicles (b). These phenotypes were observed at least three times independently with similar results. Scale bars, 1 mm (a) and 1 cm (b).
Extended Data Fig. 8 Subcellular localization of OsINP1.
Subcellular localization of OsINP1-eGFP in tobacco leaf epidermal cells. This subcellular localization was observed three times independently with similar results. Scale bars are indicated.
Extended Data Fig. 9 OsINP1 modifies plasma membrane at aperture sites.
Confocal images of tetrads stained by Calcofluor White (blue, callose wall) and CellMask Deep Red (red, membranous structures) from WT (a), osdaf1 (b), osinp1 (c) and OsINP1-eYFP transgenic plants (d). Merged fluorescent signal from YFP (yellow), Calcofluor White and CellMask Deep Red were shown in d. Arrows indicate the protruded membrane regions. These observations were repeated at least three times independently with similar results. Scale bars, 5 μm.
Extended Data Fig. 10 Signal peptide of OsDAF1 may be essential for protein stability.
a, Expression of full length OsDAF1 (upper panel), 5 amino acid truncated OsDAF1-GFP (middle panel, described in Fig. 5a) or signal peptide truncated OsDAF1-eYFP (lower panel) in tobacco leaf epidermal cells. b, The protein level of full length or truncated OsDAF1 in indicated samples, as determined by western blot. “Empty” represents a tobacco sample without Agrobacterium infiltration. Observations and data were repeated three times independently with similar results. Scale bars are indicated.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, and Supplementary Table 1.
Source data
Source Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Zhang, X., Zhao, G., Tan, Q. et al. Rice pollen aperture formation is regulated by the interplay between OsINP1 and OsDAF1. Nat. Plants 6, 394–403 (2020). https://doi.org/10.1038/s41477-020-0630-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0630-6
This article is cited by
-
Cytological Analysis and Fine Mapping of paa1 (Post-meiosis Abnormal Anther 1) Mutant with Abnormal Tapetum and Microspore Development
Biochemical Genetics (2022)
-
Grain dispersal mechanism in cereals arose from a genome duplication followed by changes in spatial expression of genes involved in pollen development
Theoretical and Applied Genetics (2022)
-
A species-specific functional module controls formation of pollen apertures
Nature Plants (2021)
-
Building portals in pollen
Nature Plants (2020)