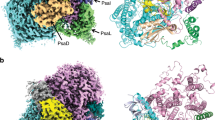
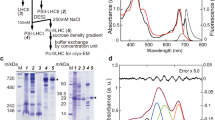
Abstract
Certain cyanobacteria synthesize chlorophyll molecules (Chl d and Chl f) that absorb in the far-red region of the solar spectrum, thereby extending the spectral range of photosynthetically active radiation1,2. The synthesis and introduction of these far-red chlorophylls into the photosynthetic apparatus of plants might improve the efficiency of oxygenic photosynthesis, especially in far-red enriched environments, such as in the lower regions of the canopy3. Production of Chl f requires the ChlF subunit, also known as PsbA4 (ref. 4) or super-rogue D1 (ref. 5), a paralogue of the D1 subunit of photosystem II (PSII) which, together with D2, bind cofactors involved in the light-driven oxidation of water. Current ideas suggest that ChlF oxidizes Chl a to Chl f in a homodimeric ChlF reaction centre (RC) complex and represents a missing link in the evolution of the heterodimeric D1/D2 RC of PSII (refs. 4,6). However, unambiguous biochemical support for this proposal is lacking. Here, we show that ChlF can substitute for D1 to form modified PSII complexes capable of producing Chl f. Remarkably, mutation of just two residues in D1 converts oxygen-evolving PSII into a Chl f synthase. Overall, we have identified a new class of PSII complex, which we term ‘super-rogue’ PSII, with an unexpected role in pigment biosynthesis rather than water oxidation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in the published article and its Supplementary Information.
Change history
03 April 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41477-020-0649-8
References
Miyashita, H. et al. Chlorophyll d as a major pigment. Nature 383, 402–402 (1996).
Chen, M., Li, Y., Birch, D. & Willows, R. D. A cyanobacterium that contains chlorophyll f —a red-absorbing photopigment. FEBS Lett. 586, 3249–3254 (2012).
Blankenship, R. E. & Chen, M. Spectral expansion and antenna reduction can enhance photosynthesis for energy production. Curr. Opin. Chem. Biol. 17, 457–461 (2013).
Ho, M.-Y., Shen, G., Canniffe, D. P., Zhao, C. & Bryant, D. A. Light-dependent chlorophyll f synthase is a highly divergent paralog of PsbA of photosystem II. Science 353, aaf9178 (2016).
Murray, J. W. Sequence variation at the oxygen-evolving centre of photosystem II: a new class of ‘rogue’ cyanobacterial D1 proteins. Photosynth. Res. 110, 177–184 (2012).
Shen, G. et al. Characterization of chlorophyll f synthase heterologously produced in Synechococcus sp. PCC 7002. Photosynth. Res. 140, 77–92 (2019).
Tilman, D., Balzer, C., Hill, J. & Befort, B. L. Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. USA 108, 20260–20264 (2011).
Cardona, T., Shao, S. & Nixon, P. J. Enhancing photosynthesis in plants: the light reactions. Essays Biochem. 62, 85–94 (2018).
Long, S. P., Marshall-Colon, A. & Zhu, X.-G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66 (2015).
Nowaczyk, M. M. et al. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of Photosystem II. Plant Cell 18, 3121–3131 (2006).
Barber, J. Photosystem II: the water splitting enzyme of photosynthesis and the origin of oxygen in our atmosphere. Q. Rev. Biophys. 49, e14 (2016).
Shen, G., Boussiba, S. & Vermaas, W. F. J. Synechocystis sp. PCC 6803 strains lacking Photosystem I and phycobilisome function. Plant Cell 5, 1853–1863 (2007).
Bečková, M. et al. Association of Psb28 and Psb27 proteins with PSII–PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol. Plant 10, 62–72 (2017).
Komenda, J. et al. Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J. Biol. Chem. 47, 48620–48629 (2004).
Umena, Y., Kawakami, K., Shen, J. R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9Å. Nature 473, 55–60 (2011).
Garg, H., Loughlin, P. C., Willows, R. D. & Chen, M. The C21-formyl group in chlorophyll f originates from molecular oxygen. J. Biol. Chem. 292, 19279–19289 (2017).
Vass, I. Molecular mechanisms of photodamage in the Photosystem II complex. Biochim. Biophys. Acta 1817, 209–217 (2012).
Masuda, T. et al. Diel regulation of photosynthetic activity in the oceanic unicellular diazotrophic cyanobacterium Crocosphaera watsonii WH8501. Environ. Microbiol. 20, 546–560 (2018).
Wegener, K. M., Nagarajan, A. & Pakrasi, H. B. An atypical psbA gene encodes a sentinel D1 protein to form a physiologically relevant inactive photosystem II complex in cyanobacteria. J. Biol. Chem. 290, 3764–3774 (2015).
Gan, F. et al. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317 (2014).
Nürnberg, D. J. et al. Photochemistry beyond the red-limit in chlorophyll f-containing systems. Science 360, 1210–1213 (2018).
Tichý, M. et al. Strain of Synechocystis PCC 6803 with aberrant assembly of photosystem II contains tandem duplication of a large chromosomal region. Front. Plant Sci. 7, 648 (2016).
Forsman, J. A., Winter, R. T. & Eaton-Rye, J. J. An improved system for the targeted mutagenesis of the psbA2 gene in Synechocystis sp. PCC 6803: mutation of D1-Glu244 to His impairs electron transfer between QA and QB of photosystem II. New Zeal. J. Bot. 57, 125–136 (2019).
Debus, R. J. et al. Does histidine 332 of the D1 polypeptide ligate the manganese cluster in photosystem II? An electron spin echo envelope modulation study. Biochemistry 40, 3690–3699 (2001).
Debus, R. J., Barry, B. A., Sithole, I., Babcock, G. T. & Mcintosh, L. Directed mutagenesis indicates that the donor to P680+ in photosystem II is tyrosine-161 of the D1 polypeptide. Biochemistry 27, 9071–9074 (1988).
Ried, J. L. & Collmer, A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange–eviction mutagenesis. Gene 57, 239–246 (1987).
Williams, J. G. K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167, 766–778 (1988).
Hollingshead, S. et al. Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287, 27823–27833 (2012).
Zhu, B., Cai, G., Hall, E. O. & Freeman, G. J. In-FusionTM assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques 43, 354–359 (2007).
Komenda, J. & Barber, J. Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34, 9625–9631 (1995).
Suzuki, H. et al. Functional roles of D2-Lys317 and the interacting chloride ion in the water oxidation reaction of photosystem II as revealed by Fourier transform infrared analysis. Biochemistry 52, 4748–4757 (2013).
Wittig, I., Karas, M. & Schägger, H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteom. 6, 1215–1225 (2007).
Dobáková, M., Sobotka, R., Tichý, M. & Komenda, J. Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149, 1076–1086 (2009).
Komenda, J., Tichý, M. & Eichacker, L. A. The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 46, 1477–1483 (2005).
Rögner, M., Nixon, P. J. & Diner, B. A. Purification and characterization of photosystem I and photosystem II core complexes from wild-type and phycocyanin-deficient strains of the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 265, 6189–6196 (1990).
Komenda, J. et al. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 22390–22399 (2008).
Bumba, L., Prasil, O. & Vacha, F. Antenna ring around trimeric Photosystem I in chlorophyll b containing cyanobacterium Prochlorothrix hollandica. Biochim. Biophys. Acta Bioenerg. 1708, 1–5 (2005).
Li, N. et al. PsaD is required for the stable binding of PsaC to the photosystem I core protein of Synechococcus sp. PCC 6301. Biochemistry 30, 7863–7872 (1991).
Dobáková, M., Tichý, M. & Komenda, J. Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 145, 1681–1691 (2007).
Yu, J. et al. Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein. Proc. Natl Acad. Sci. USA 115, E7824–E7833 (2018).
Komenda, J. et al. The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 158, 476–486 (2012).
Bučinská, L. et al. The ribosome-bound protein Pam68 promotes insertion of chlorophyll into the CP47 subunit of photosystem II. Plant Physiol. 176, 2931–2942 (2018).
Ritchie, R. J. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth. Res. 89, 27–41 (2006).
Li, Y., Scales, N., Blankenship, R. E., Willows, R. D. & Chen, M. Extinction coefficient for red-shifted chlorophylls: chlorophyll d and chlorophyll f. Biochim. Biophys. Acta Bioenerg. 1817, 1292–1298 (2012).
Acknowledgements
We thank B. Nwaobi for excellent laboratory management and R. Debus and J. Eaton-Rye for providing plasmids and strains. Financial support was provided by the Biotechnology and Biological Sciences Research Council (BB/P00931X/1 to J.W.M. and P.J.N.), the Grant Agency of the Czech Republic (project no. 19-29225X to M.B., R.S. and J.K.) and the Ministry of Education, Youth and Sports of the Czech Republic (National Program of Sustainability I, ID: LO1416 to M.B., R.S. and J.K.). J.P.T. received a PhD scholarship from the Indonesia Endowment Fund for Education (LPDP).
Author information
Authors and Affiliations
Contributions
P.J.N., J.K. and J.W.M. conceived the research. J.P.T., S.S and J.K. prepared the figures. P.J.N and J.K. wrote the manuscript with contributions from all the other authors. P.J.N. and J.K. coordinated the activities. J.P.T. constructed the mutants. J.P.T., M.B., S.S., J.Y., Z.Z. and R.S. performed the biochemical analyses. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of FLAG-ChlF complexes immunopurified from the FLAG-ChlF/WT strain.
a, FLAG-ChlF complexes isolated from the FLAG-ChlF/WT strain were separated by clear-native polyacrylamide gel electrophoresis (CN-PAGE) in the first dimension (1D colour) and visualized by fluorescence (1D fluor) then denatured, separated by SDS-PAGE in the second dimension and protein subunits stained by Sypro Orange (2D SYPRO). Proteins were identified in the stained gel by immunoblotting and mass spectrometry. Abbreviations: F.ChlF, FLAG-tagged ChlF; PSII(1)’, monomeric PSII-like complex containing ChlF; RC47’, PSII(1)’ complex lacking CP43 and containing ChlF; PSI(1)-RC47’, complex composed of PSI monomer and RC47’ complex; CP43, CP43 module; U.P., unassembled proteins. b, Cartoon representations comparing the monomeric PSII complex containing D1 (PSII(1)) and the PSII-like complexes containing ChlF (PSII(1)’ and RC47’). Small subunits are not shown for clarity.
Extended Data Fig. 2 Characterization of FLAG-ChlF complexes immunopurified from the FLAG-ChlF/ΔD1/ΔCP43 strain.
a, Cartoon showing the psbD1C locus found in the FLAG-ChlF/ΔD1strain and the resulting FLAG-ChlF/ΔD1/ΔCP43 strain following deletion of psbD1C. Positions of the forward (F4) and reverse (R4) primers (Supplementary Table 3) used for PCR genotyping and predicted sizes are indicated. b, Agarose gel of PCR fragments confirming the genotype of the FLAG-ChlF/ΔD1/ΔCP43 strain and non-tagged strains. c, FLAG-ChlF complexes isolated from the FLAG-ChlF/ΔD1/ΔCP43 strain were separated by clear-native polyacrylamide gel electrophoresis (CN-PAGE) in the first dimension (1D colour) and visualized by fluorescence (1D fluor) then denatured, separated by SDS-PAGE in the second dimension and protein subunits stained by Sypro Orange (2D SYPRO). Proteins were identified in the stained gel by immunoblotting (FLAG and D2 blots shown) and mass spectrometry. Abbreviations: F.ChlF, FLAG-tagged ChlF; RC47’, PSII(1)’ complex lacking CP43; PSI(1)-RC47’, complex composed of PSI monomer and RC47’ complex; U.P., unassembled proteins. d, Cartoon showing the RC47’ complex composed of ChlF/D2 heterodimer with attached CP47. Small subunits are not shown for clarity.
Extended Data Fig. 3 Characterization of FLAG-ChlF complexes immunopurified from the FLAG-ChlF/ΔD1/ΔCP47 strain.
a, Cartoon showing the psbB locus found in the FLAG-ChlF/ΔD1strain and the resulting FLAG-ChlF/ΔD1/ΔCP47 strain following deletion of psbB. Positions of the forward (F6) and reverse (R6) primers (Supplementary Table 3) used for PCR genotyping and predicted sizes are indicated. b, Agarose gel of PCR fragments confirming the genotype of the FLAG-ChlF/ΔD1/ΔCP47 strain and untagged strains. c, FLAG-ChlF complexes isolated from the FLAG-ChlF/ΔD1/ΔCP47 strain were separated by clear-native polyacrylamide gel electrophoresis (CN-PAGE) in the first dimension (1D colour) and visualized by fluorescence (1D fluor) then denatured, separated by SDS-PAGE in the second dimension and protein subunits stained by Sypro Orange (2D SYPRO). Proteins were identified in the stained gel by immunoblotting (D2 and FLAG blots shown) and mass spectrometry. Abbreviations: F.ChlF, FLAG-tagged ChlF; PSI(3), trimeric PSI; PSI(1)-RCa’, complex composed of PSI monomer and ChlF/D2 complex; RC43’, complex composed of ChlF/D2 with CP43 attached; RCIIa’, assembly complex of ChlF/D2 with accessory proteins; RCIIb’, assembly complex of ChlF/D2 lacking assembly factors. U.P., unassembled proteins. d, Cartoon showing the modified RC43’ complex, composed of a ChlF/D2 heterodimer with attached CP43 and the RCIIb’ complex. Small subunits are not shown for clarity.
Extended Data Fig. 4 Detection of FLAG-ChlF and FLAG-D1.
Detergent-solubilized membrane proteins from the a, FLAG-ChlF/PSII−strain or b, FLAG-D1/PSII− strain were separated by clear-native (CN) gel electrophoresis in the first dimension (1D colour) and visualized by fluorescence (1D fluor) then denatured, separated by SDS-PAGE in the second dimension and protein subunits stained by Sypro Orange (2D SYPRO), then transferred to PVDF membranes for immunochemical detection of FLAG-tagged proteins using specific FLAG-tag antibodies (FLAG). Abbreviations: PSI(3), trimeric PSI; PSI(1), monomeric PSI complex; U.P., unassembled proteins.
Extended Data Fig. 5 Analysis of FLAG-ChlF complexes in the FLAG-ChlF(MG)/ΔD1 mutant (ChlF-MG) in which the QD pair (Q149/D150) of ChlF is replaced by the MG pair found in D1.
Detergent-solubilized membrane proteins from the FLAG-ChlF(MG)/ΔD1 strain were separated by clear-native polyacrylamide gel electrophoresis (CN-PAGE) in the first dimension (1D colour) and visualized by fluorescence (1D fluor) then denatured, separated by SDS-PAGE in the second dimension and protein subunits stained by Sypro Orange (2D SYPRO), then transferred to PVDF membrane for immunochemical detection of FLAG-ChlF (FLAG), D2, CP47 and CP43 proteins. Abbreviations: PSI(3), trimeric PSI; PSII(1), monomeric PSII; PSI(1), monomeric PSI complex; U.P., unassembled proteins.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Tables 1–3.
Supplementary Data
Unprocessed Western Blot Supplementary Fig. 4.
Source data
Source Data Extended Data Fig. 2
Unprocessed western blot Extended Data Fig. 2c.
Source Data Extended Data Fig. 3
Unprocessed western blot Extended Data Fig. 3c.
Source Data Extended Data Fig. 4
Unprocessed Wwestern blot Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Unprocessed western blot Extended Data Fig. 5.
Rights and permissions
About this article
Cite this article
Trinugroho, J.P., Bečková, M., Shao, S. et al. Chlorophyll f synthesis by a super-rogue photosystem II complex. Nat. Plants 6, 238–244 (2020). https://doi.org/10.1038/s41477-020-0616-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0616-4