Abstract
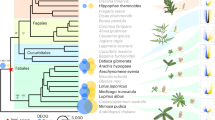
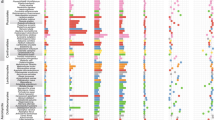
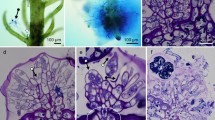
Plants are the foundation of terrestrial ecosystems, and their colonization of land was probably facilitated by mutualistic associations with arbuscular mycorrhizal fungi. Following this founding event, plant diversification has led to the emergence of a tremendous diversity of mutualistic symbioses with microorganisms, ranging from extracellular associations to the most intimate intracellular associations, where fungal or bacterial symbionts are hosted inside plant cells. Here, through analysis of 271 transcriptomes and 116 plant genomes spanning the entire land-plant diversity, we demonstrate that a common symbiosis signalling pathway co-evolved with intracellular endosymbioses, from the ancestral arbuscular mycorrhiza to the more recent ericoid and orchid mycorrhizae in angiosperms and ericoid-like associations of bryophytes. By contrast, species forming exclusively extracellular symbioses, such as ectomycorrhizae, and those forming associations with cyanobacteria, have lost this signalling pathway. This work unifies intracellular symbioses, revealing conservation in their evolution across 450 million yr of plant diversification.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All assemblies and gene annotations generated in this project can be found in SymDB (www.polebio.lrsv.ups-tlse.fr/symdb/). Raw sequencing data can be found under NCBI Bioproject PRJNA576233 (B. pusilla), PRJNA362997 and PRJNA362995 (M. paleacea genome and transcriptome, respectively) and PRJNA576577 (M. polymorpha ssp. montivagans and ssp. polymorpha).
References
Gensel, P. G. The Emerald Planet: How Plants Changed Earth’s History (Oxford Univ. Press, 2008).
Parniske, M. Arbuscular mycorrhizae: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775 (2008).
Delaux, P. M., Séjalon-Delmas, N., Bécard, G. & Ané, J. M. Evolution of the plant–microbe symbiotic ‘toolkit’. Trends Plant Sci. 18, 298–304 (2013).
Werner, G. D. A. et al. Symbiont switching and alternative resource acquisition strategies drive mutualism breakdown. Proc. Natl Acad. Sci. USA 115, 5229–5234 (2018).
Smith, S. & Read, D. Mycorrhizal Symbiosis (Academic Press, 2008).
Kottke, I. et al. Heterobasidiomycetes form symbiotic associations with hepatics: Jungermanniales have sebacinoid mycobionts while Aneura pinguis (Metzgeriales) is associated with a Tulasnella species. Mycol. Res. 107, 957–968 (2003).
Griesmann, M. et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361, eaat1743 (2018).
Wang, B. & Qiu, Y. L. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363 (2006).
Delaux, P. M., Radhakrishnan, G. & Oldroyd, G. Tracing the evolutionary path to nitrogen-fixing crops. Curr. Opin. Plant Biol. 26, 95–99 (2015).
Martin, F. M., Uroz, S. & Barker, D. G. Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356, eaad4501 (2017).
van Velzen, R. et al. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl Acad. Sci. USA 115, E4700–E4709 (2018).
Albalat, R. & Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 17, 379–391 (2016).
Tabach, Y. et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature 493, 694–698 (2013).
Delaux, P. M. Comparative phylogenomics of symbiotic associations. New Phytol. 213, 89–94 (2017).
Dey, G., Jaimovich, A., Collins, S. R., Seki, A. & Meyer, T. Systematic discovery of human gene function and principles of modular organization through phylogenetic profiling. Cell Rep. 10, 993–1006 (2015).
Bravo, A., York, T., Pumplin, N., Mueller, L. A. & Harrison, M. J. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants 2, 15208 (2016).
Delaux, P. M. et al. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 10, e1004487 (2014).
Favre, P. et al. A novel bioinformatics pipeline to discover genes related to arbuscular mycorrhizal symbiosis based on their evolutionary conservation pattern among higher plants. BMC Plant Biol. 14, 333 (2014).
Xue, L. et al. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 167, 854–871 (2015).
Keymer, A. et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6, e29107 (2017).
Bravo, A., Brands, M., Wewer, V., Dörmann, P. & Harrison, M. J. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 214, 1631–1645 (2017).
Grosche, C., Genau, A. C. & Rensing, S. A. Evolution of the symbiosis-specific GRAS regulatory network in bryophytes. Front. Plant Sci. 9, 1621 (2018).
Delaux, P.-M. et al. Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl Acad. Sci. USA 112, 13390–13395 (2015).
Wang, B. et al. Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186, 514–525 (2010).
Humphreys, C. P. et al. Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat. Commun. 1, 103 (2010).
Bowman, J. L. et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304 (2017).
Brundrett, M. & Tedersoo, L. Misdiagnosis of mycorrhizas and inappropriate recycling of data can lead to false conclusions. New Phytol. 221, 18–24 (2019).
Villarreal A, J. C., Crandall-Stotler, B. J., Hart, M. L., Long, D. G. & Forrest, L. L. Divergence times and the evolution of morphological complexity in an early land plant lineage (Marchantiopsida) with a slow molecular rate. New Phytol. 209, 1734–1746 (2016).
Read, D. J. & Perez-Moreno, J. Mycorrhizas and nutrient cycling in ecosystems—a journey towards relevance? New Phytol. 157, 475–492 (2003).
Rey, T. et al. The Medicago truncatula GRAS protein RAD1 supports arbuscular mycorrhiza symbiosis and Phytophthora palmivora susceptibility. J. Exp. Bot. 68, 5871–5881 (2017).
Park, H.-J., Floss, D. S., Levesque-Tremblay, V., Bravo, A. & Harrison, M. J. Hyphal branching during arbuscule development requires RAM1. Plant Physiol. 169, 2774–2788 (2015).
Luginbuehl, L. H. et al. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178 (2017).
Jiang, Y. et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175 (2017).
Adams, D. G. The Ecology of cyanobacteria: their diversity in time and space (Kluwer, 2000); https://doi.org/10.1016/0303-2647(92)90025-T
Li, F. W. et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants 4, 460–472 (2018).
Cope, K. R. et al. The ectomycorrhizal fungus Laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway to colonize Populus roots. Plant Cell 31, 2386–2410 (2019).
Miura, C. et al. The mycoheterotrophic symbiosis between orchids and mycorrhizal fungi possesses major components shared with mutualistic plant–mycorrhizal symbioses. Mol. Plant Microbe Interact. 31, 1032–1047 (2018).
Oba, H., Tawaray, K. & Wagatsuma, T. Arbuscular mycorrhizal colonization in Lupinus and related genera. Soil Sci. Plant Nutr. 47, 685–694 (2001).
Oldroyd, G. E. D. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263 (2013).
Gutjahr, C. et al. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20, 2989–3005 (2008).
Gherbi, H. et al. SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankiabacteria. Proc. Natl Acad. Sci. USA 105, 4928–4932 (2008).
Svistoonoff, S. et al. The independent acquisition of plant root nitrogen-fixing symbiosis in fabids recruited the same genetic pathway for nodule organogenesis. PLoS ONE 8, e64515 (2013).
Buendia, L., Wang, T., Girardin, A. & Lefebvre, B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 210, 184–195 (2016).
Capoen, W. et al. Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell 21, 1526–1540 (2009).
Gleason, C. et al. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152 (2006).
Singh, S., Katzer, K., Lambert, J., Cerri, M. & Parniske, M. CYCLOPS, A DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152 (2014).
Jin, Y. et al. IPD3 and IPD3L function redundantly in rhizobial and mycorrhizal symbioses. Front. Plant Sci. 9, 267 (2018).
Yasumura, Y., Crumpton-Taylor, M., Fuentes, S. & Harberd, N. P. Step-by-step acquisition of the gibberellin–DELLA growth-regulatory mechanism during land-plant evolution. Curr. Biol. 17, 1225–1230 (2007).
Soltis, P. S. & Soltis, D. E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30, 159–165 (2016).
Valdés-López, O. et al. A novel positive regulator of the early stages of root nodule symbiosis identified by phosphoproteomics. Plant Cell Physiol. 60, 575–586 (2019).
Murray, J. D. et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65, 244–252 (2011).
Imaizumi-Anraku, H. et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433, 527–531 (2005).
Liu, C. W. et al. A protein complex required for polar growth of rhizobial infection threads. Nat. Commun. 10, 2848 (2019).
Pumplin, N. et al. Medicago truncatula vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61, 482–494 (2010).
Feddermann, N. et al. The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J. 64, 470–481 (2010).
Takeda, N., Tsuzuki, S., Suzaki, T., Parniske, M. & Kawaguchi, M. CERBERUS and NSP1 of Lotus japonicus are common symbiosis genes that modulate arbuscular mycorrhiza development. Plant Cell Physiol. 54, 1711–1723 (2013).
Huisman, R. et al. A symbiosis-dedicated SYNTAXIN OF PLANTS 13II isoform controls the formation of a stable host–microbe interface in symbiosis. New Phytol. 211, 1338–1351 (2016).
Healey, A., Furtado, A., Cooper, T. & Henry, R. J. A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10, 1–8 (2014).
Luo, R. et al. Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 4, 30 (2015).
Leggett, R. M., Clavijo, B. J., Clissold, L., Clark, M. D. & Caccamo, M. Next clip: an analysis and read preparation tool for nextera long mate pair libraries. Bioinformatics 30, 566–568 (2014).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Roberts, R. J., Carneiro, M. O. & Schatz, M. C. The advantages of SMRT sequencing. Genome Biol. 14, 405 (2013).
Chin, C. S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Parra, G., Bradnam, K. & Korf, I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007).
Acknowledgements
This work was supported by the Agence Nationale de la Recherche (ANR) grant EVOLSYM (ANR-17-CE20-0006-01) to P.-M.D., by the Bill and Melinda Gates Foundation as Engineering the Nitrogen Symbiosis for Africa (OPP1172165), by the BBSRC as OpenPlant to G.E.D.O (BB/L014130/1), by the 10KP initiative (BGI-Shenzhen), by the National Science Foundation (DEB1831428) to F.-W.L. and by the Swedish Research Council Vetenskapsrådet (VR) to U.L. (2011‐5609 and 2014‐522) and to D.M.E (2016-05180). G.V.R is additionally supported by a Biotechnology and Biological Sciences Research Council Discovery Fellowship (BB/S011005/1). Part of this work was conducted at the Laboratoire de Recherche en Sciences Végétales (LRSV) laboratory, which belongs to the TULIP Laboratoire d’Excellence (ANR-10-LABX-41). We are grateful to the genotoul bioinformatics platform Toulouse Midi–Pyrenees for providing computing and storage resources. We thank F. Roux for helping with the collection of M. polymorpha accessions, A. Cooke for assistance with M. paleacea DNA extraction, P. Szoevenyi for advice on M. polymorpha DNA extraction, and D. Barker and members of the Engineering Nitrogen Symbiosis for Africa (ENSA) project for helpful comments and discussion. Figure 6b was prepared by J. Calli (www.jeremy-calli.fr).
Author information
Authors and Affiliations
Contributions
P.-M.D., G.V.R., M.K.R., J.K., G.E.D.O. and T.V. conceived the experiments; J.K., H.S.C. and L.C. developed symDB; G.V.R., M.K.R., J.K., T.V., D.L.M.M., N.V., C.L., J.C. and P.-M.D. conducted the experiments; A.-M.L., D.M.E. and U.L. generated the M. polymorpha subspecies genomes; F.-W.L., S.C. and G.K.S.W. generated the B. pusilla transcriptome; P.-M.D., G.V.R., M.K.R., J.K. and T.V. analysed the data; J.K. compiled the supplementary material; G.V.R., M.K.R., G.E.D.O. and P.-M.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Andrea Genre and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods and Supplementary Figs. 1–37.
Supplementary Tables
Supplementary Tables 1–7, including title and description for each table.
Rights and permissions
About this article
Cite this article
Radhakrishnan, G.V., Keller, J., Rich, M.K. et al. An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants 6, 280–289 (2020). https://doi.org/10.1038/s41477-020-0613-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0613-7
This article is cited by
-
Genome sequencing and functional analysis of a multipurpose medicinal herb Tinospora cordifolia (Giloy)
Scientific Reports (2024)
-
IPD3, a master regulator of arbuscular mycorrhizal symbiosis, affects genes for immunity and metabolism of non-host Arabidopsis when restored long after its evolutionary loss
Plant Molecular Biology (2024)
-
A rare non-canonical splice site in Trema orientalis SYMRK does not affect its dual symbiotic functioning in endomycorrhiza and rhizobium nodulation
BMC Plant Biology (2023)
-
Mycorrhizas drive the evolution of plant adaptation to drought
Communications Biology (2023)
-
Comparative phylotranscriptomics reveals ancestral and derived root nodule symbiosis programmes
Nature Plants (2023)