Abstract
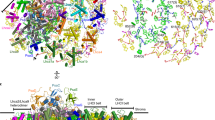
Solar energy harnessed by oxygenic photosynthesis supports most of the life forms on Earth. In eukaryotes, photosynthesis occurs in chloroplasts and is achieved by membrane-embedded macromolecular complexes that contain core and peripheral antennae with multiple pigments. The structure of photosystem I (PSI) comprises the core and light-harvesting (LHCI) complexes, which together form PSI–LHCI. Here we determined the structure of PSI–LHCI from the salt-tolerant green alga Dunaliella salina using X-ray crystallography and electron cryo-microscopy. Our results reveal a previously undescribed configuration of the PSI core. It is composed of only 7 subunits, compared with 14–16 subunits in plants and the alga Chlamydomonas reinhardtii, and forms the smallest known PSI. The LHCI is poorly conserved at the sequence level and binds to pigments that form new energy pathways, and the interactions between the individual Lhca1–4 proteins are weakened. Overall, the data indicate the PSI of D. salina represents a different type of the molecular organization that provides important information for reconstructing the plasticity and evolution of PSI.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this Article and its Extended data and Supplementary tables. The cryo-EM map has been deposited into the Electron Microscopy Data Bank with accession code EMD-4883. The atomic models have been deposited in the PDB under accession codes 6QPH and 6RHZ.
References
Iwai, M., Grob, P., Iavarone, A. T., Nogales, E. & Niyogi, K. K. A unique supramolecular organization of photosystem I in the moss Physcomitrella patens. Nat. Plants 4, 904–909 (2018).
Pi, X. et al. Unique organization of photosystem I–light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc. Natl Acad. Sci. USA 115, 4423–4428 (2018).
Pinnola, A. et al. A LHCB9-dependent photosystem I megacomplex induced under low light in Physcomitrella patens. Nat. Plants 4, 910–919 (2018).
Amunts et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Kühlbrandt, W. The resolution revolution. Science 343, 1443–1444 (2014).
Wei, X. et al. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 534, 69–74 (2016).
Su, X. et al. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 357, 815–820 (2017).
Hahn, A., Vonck, J., Mills, D. J., Meier, T. & Kühlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360, eaat4318 (2018).
Pan, X. et al. Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II. Science 360, 1109–1113 (2018).
Perez Boerema, A. et al. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 4, 212–217 (2018).
Su, X. et al. Antenna arrangement and energy transfer pathways of a green algal photosystem-I–LHCI supercomplex. Nat. Plants 5, 273–281 (2019).
Qin, X. et al. Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits. Nat. Plants 5, 263–272 (2019).
Qin, X., Suga, M., Kuang, T. & Shen, J. R. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348, 989–995 (2015).
Mazor, Y., Borovikova, A., Caspy, I. & Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 3, 17014 (2017).
Kim, M., Park, S., Polle, J. E. & Jin, E. Gene expression profiling of Dunaliella sp. acclimated to different salinities. Phycol. Res. 58, 17–28 (2010).
Xu, H. et al. Light intensity dependent expression of Lhca gene family encoding LHCI in Dunaliella salina. Afr. J. Microbiol. Res. 6, 6165–6171 (2012).
Smith, B. M. et al. Response of the photosynthetic apparatus in Dunaliella salina (green algae) to irradiance stress. Plant Physiol. 93, 1433–1440 (1990).
Varsano, T., Wolf, S. G. & Pick, U. A chlorophyll a/b-binding protein homolog that is induced by iron deficiency is associated with enlarged photosystem I units in the eukaryotic alga Dunaliella salina. J. Biol. Chem. 281, 10305–10315 (2006).
Saha, S. K., Kazipet, N. & Murray, P. The carotenogenic Dunaliella salina CCAP 19/20 produces enhanced levels of carotenoid under specific nutrients limitation. BioMed Res. Int. 2018, 7532897 (2018).
Raja, R., Hemaiswarya, S. & Rengasamy, R. Exploitation of Dunaliella for β-carotene production. Appl. Microbiol. Biotechnol. 74, 517–523 (2007).
Lamers, P. P., Janssen, M., De Vos, R. C., Bino, R. J. & Wijffels, R. H. Exploring and exploiting carotenoid accumulation in Dunaliella salina for cell-factory applications. Trends Biotechnol. 26, 631–638 (2008).
Slocombe, S. P. et al. Unlocking nature’s treasure-chest: screening for oleaginous algae. Sci. Rep. 5, 9844 (2015).
Amunts, A., Toporik, H., Borovikova, A. & Nelson, N. Structure determination and improved model of plant photosystem I. J. Biol. Chem. 285, 3478–3486 (2010).
Amunts, A. & Nelson, N. Plant photosystem I design in the light of evolution. Structure 17, 637–650 (2009).
Polle, J. et al. Draft nuclear genome sequence of the halophilic and beta-carotene-accumulating green alga Dunaliella salina strain CCAP19/18. Genome Announc. 5, e01105-17 (2017).
Ozawa, S. I. et al. Configuration of ten light-harvesting chlorophyll a/b complex I subunits in Chlamydomonas reinhardtii photosystem I. Plant Physiol. 178, 583–595 (2018).
Malavath, T., Caspy, I., Netzer-El, S. Y., Klaiman, D. & Nelson, N. Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochim. Biophys. Acta 1859, 645–654 (2018).
Jensen, P. E., Haldrup, A. H., Rosgaard, L. & Scheller, H. V. Molecular dissection of photosystem I in higher plants: topology, structure and function. Physiol. Plant. 119, 313–321 (2003).
Ozawa, S. I., Onishi, T. & Takahashi, Y. Identification and characterization of an assembly intermediate subcomplex of photosystem I in the green alga Chlamydomonas reinhardtii. J. Biol. Chem. 285, 20072–20079 (2010).
Li, Y. et al. Transcriptome analysis reveals regulation of gene expressionduring photoacclimation to high irradiance levels in Dunaliella salina (Chlorophyceae). Phycol. Res. 67, 291–302 (2019).
Petrou, K. et al. State transitions and nonphotochemical quenching during a nutrient-induced fluorescence transient in phosphorus-starved Dunaliela tertiolecta. J. Phycol. 44, 1204–1211 (2008).
Safie, S. R. B. Responses to Changes in Light Intensity and Salinity in Microalga Dunaliella tertiolecta. PhD thesis, National Univ. Singapore (2017).
Mazor, Y., Nataf, D., Toporik, H. & Nelson, N. Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803. eLife 3, e01496 (2014).
Wientjes, E., van Stokkum, I. H., van Amerongen, H. & Croce, R. The role of the individual Lhcas in photosystem I excitation energy trapping. Biophys. J. 101, 745–754 (2011).
Suga, M., Qin, X., Kuang, T. & Shen, J. R. Structure and energy transfer pathways of the plant photosystem I-LHCI supercomplex. Curr. Opin. Struct. Biol. 39, 46–53 (2016).
Suga, M. et al. Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I. Nat. Plants 5, 626–636 (2019).
Hong, L., Liu, J. L., Midoun, S. Z. & Miller, P. C. Transcriptome sequencing and annotation of the halophytic microalga Dunaliella salina. J. Zhejiang Univ. Sci. B 18, 833–844 (2017).
Wei, S. et al. Salinity-induced palmella formation mechanism in halotolerant algae Dunaliella salina revealed by quantitative proteomics and phosphoproteomics. Front. Plant Sci. 8, 810 (2017).
Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H., Núñez-Ramírez, R., Sorzano, C. O., Carazo, J. M. & Marabini, R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3, 977–990 (2008).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
De la Rosa-Trevín, J. M. et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1993).
Roth, M. J., Tanese, N. & Goff, S. P. Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J. Biol. Chem. 260, 9326–9335 (1985).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. J. Basic local alignment search tool. Mol. Biol. 215, 403–410 (1990).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D 66, 22–25 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
DeLano, W. L. Pymol: An open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 40, 82–92 (2002).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2008).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
The cryo-EM data were collected at the ESRF beamline CM01, experiment MX-2090. We thank G. Effantin for the outstanding support with the grid screening and data collection; the staff scientists of the SciLifeLab cryo-EM facility, funded by the Knut and Alice Wallenberg, Erling-Persson Family and Kempe foundations. The X-ray diffraction data were collected at the SLS and ESRF. We thank the staff scientists for excellent guidance and assistance; A. Kaplan for providing the green alga Chlorella ohadii. This work was supported by the Israel Science Foundation (569/17), Joint UGC-ISF (2716/17), German-Israeli Foundation for Scientific Research and Development (1483), Swedish Foundation for Strategic Research (FFL15:0325), Ragnar Söderberg Foundation (M44/16), Swedish Research Council (NT_2015-04107), European Research Council (ERC-2018-StG-805230) and Knut and Alice Wallenberg Foundation (2018.0080). A.A. is supported by the EMBO Young Investigator Program.
Author information
Authors and Affiliations
Contributions
A.P.-B., D.K., I.C., S.Y.N.-E., A.A. and N.N. performed the research. A.P.-B., A.A. and N.N. analysed the data. A.P.-B., A.A. and N.N. wrote the manuscript. All of the authors discussed and commented on the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Poul Erik Jensen, Mei Li and the other, anonymous, reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 X-ray crystal structure determination.
(a) Polypeptide composition of D. salina mini-PSI used for crystallization, and comparison with related organisms. The experiments described were independently repeated at least three times (b) activity in NADP photoreduction. (c) Crystals of PSI. Over one hundred of similar wells were generated in the crystallization plates. (d) A representative diffraction pattern showing single spots reaching ~ 3 Å resolution anisotropically.
Extended Data Fig. 2
Cryo-EM data processing workflow.
Extended Data Fig. 3 Quality of the cryo-EM map.
(a) The final map colored by local resolution and viewed from the surface and cut- through. Examples of the best (b) and worst (c) modeled densities for ligands.
Extended Data Fig. 4 Unassigned densities.
Six discrete weak densities were identified in the membrane bilayer that could not be unambiguously assigned. The densities likely represent non-protein components that do not affect the overall conclusions.
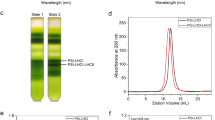
Extended Data Fig. 5 Purification profile of PSI-LHCI from D. salina thylakoids.
MW; lane 1: thylakoid membranes after solubilization; lane 2: dark-green fraction of TOYOPEARL DEAE-650C; lane 3: fraction eluted from sucrose gradient; lane 4: elution from FPLC SOURCE 15Q; lane 5: after second sucrose gradient. A subunit distribution of PSI is indicated according to MS data. The experiment was repeated at least three times.
Extended Data Fig. 6 Mini-PSI analysis.
(a) Mild solubilization of the thylakoid membrane with two detergents. Lane 1: MW, lane 2: D. salina thylakoid membranes solubilized in α-DDM, lane 3: D. salina thylakoid membranes solubilized in β-DDM. Both preparations indicate that PSI is found in two different forms. The experiment was repeated twice. (b) Comparison of the mildly solubilized D. salina PSI with purified mini-PSI indicates that the complexes correspond to each other, and small differences in migration can be explained by presence/absence of lipids. Lane 1: MW, lane 2: D. salina thylakoids membranes solubilized in β-DDM, lane 3: purified D. salina PSI-LCHI. This experiment was repeated 5 times. (c) Solubilized membranes were applied on a sucrose gradient 15 to 50% The indicated bands 1 and 2, representing two forms of PSI were run on 20% polyacrylamide urea gel for protein separation. (d) Quantification of the bands from (c). The data suggests that fraction 2 is enriched with mini-PSI, lacking some of the core subunit. The experiment was repeated 5 times with similar results.
Extended Data Fig. 7 Major energy pathways.
(a) Major energy pathways are shown with dashed lines, distances are magnesium to magnesium (Å). The newly identified potential energy pathways are shown in orange and black. (b) Names and locations of the pathways are shown in the table and the newly identified potential energy pathways are shown in orange as highlighted in A.
Extended Data Fig. 8 Identification of new carotenes in Lhca3.
Chlorophylls are shown in green, conserved carotenes are colored in dark orange, represented in yellow are the newly identified carotenes in D. salina and in dark grey is chlorophyll a313 present in plant (PDBID:4KX8) but not in D. salina.
Extended Data Fig. 10 Comparison of the structures and sequences of LHCI between green algae.
The superposition between LHCI proteins of D. salina and C. reinhardtii shows that C-terminus extensions of Lhca7 and Lhca3 responsible for the binding the outer belt are missing in D. salina.
Supplementary information
Supplementary Information
Supplementary Tables 1–7.
Rights and permissions
About this article
Cite this article
Perez-Boerema, A., Klaiman, D., Caspy, I. et al. Structure of a minimal photosystem I from the green alga Dunaliella salina. Nat. Plants 6, 321–327 (2020). https://doi.org/10.1038/s41477-020-0611-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0611-9
This article is cited by
-
Structural insights into a unique PSI–LHCI–LHCII–Lhcb9 supercomplex from moss Physcomitrium patens
Nature Plants (2023)
-
High salt-induced PSI-supercomplex is associated with high CEF and attenuation of state transitions
Photosynthesis Research (2023)
-
Spectral diversity of photosystem I from flowering plants
Photosynthesis Research (2023)
-
The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4
Nature Plants (2022)
-
Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex
Nature Plants (2022)