Abstract
Growth responses to competition1 and defence responses to the attack of consumer organisms2 are two classic examples of adaptive phenotypic plasticity in plants. However, the mechanistic and functional links between these responses are not well understood. Jasmonates, a family of lipid-derived signals, are potent growth inhibitors and central regulators of plant immunity to herbivores and pathogens3,4, with both roles being evolutionarily conserved from bryophytes5 to angiosperms6. When shade-intolerant plants perceive the proximity of competitors using the photoreceptor phytochrome B, they activate the shade-avoidance syndrome and downregulate jasmonate responses7. Despite the central implications of this light-mediated change in the growth/defence balance for plant adaptation and crop yield8,9, the mechanisms by which photoreceptors relay light cues to the jasmonate signalling pathway remain poorly understood10. Here, we identify a sulfotransferase (ST2a) that is strongly upregulated by plant proximity perceived by phytochrome B via the phytochrome B–phytochrome interacting factor signalling module. By catalysing the formation of a sulfated jasmonate derivative, ST2a acts to reduce the pool of precursors of active forms of jasmonates and represents a direct molecular link between photoreceptors and hormone signalling in plants. The metabolic step defined by this enzyme provides a molecular mechanism for prioritizing shade avoidance over defence under intense plant competition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
Schmitt, J., Stinchcombe, J. R., Heschel, M. S. & Huber, H. The adaptive evolution of plasticity: phytochrome-mediated shade avoidance responses. Integr. Comp. Biol. 43, 459–469 (2003).
Farmer, E. E. Leaf Defence (Oxford Univ. Press, 2014).
Wasternack, C. & Feussner, I. The oxylipin pathways: biochemistry and function. Annu. Rev. Plant Biol. 69, 363–386 (2018).
Howe, G. A., Major, I. T. & Koo, A. J. Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 69, 387–415 (2018).
Monte, I. et al. A single JAZ repressor controls the jasmonate pathway in Marchantia polymorpha. Mol. Plant 12, 185–198 (2019).
Guo, Q. et al. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl Acad. Sci. USA 115, E10768–E10777 (2018).
Moreno, J. E., Tao, Y., Chory, J. & Ballaré, C. L. Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl Acad. Sci. USA 106, 4935–4940 (2009).
Ballaré, C. L. & Austin, A. T. Recalculating growth and defense strategies under competition: key roles of photoreceptors and jasmonates. J. Exp. Bot. 70, 3425–3434 (2019).
Campos, M. L. et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7, 12570 (2016).
Ballaré, C. L. Light regulation of plant defense. Annu. Rev. Plant Biol. 65, 335–363 (2014).
de Wit, M., Galvão, V. C. & Fankhauser, C. Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67, 613–617 (2016).
Cerrudo, I. et al. Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158, 2042–2052 (2012).
Leone, M., Keller, M. M., Cerrudo, I. & Ballaré, C. L. To grow or defend? Low red:far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol. 204, 355–367 (2014).
Chico, J. M. et al. Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26, 1967–1980 (2014).
Liu, Y. et al. Arabidopsis FHY3 and FAR1 proteins regulate the balance between growth and defense responses under shade conditions. Plant Cell 31, 2089–2106 (2019).
Yamashino, T., Kitayama, M. & Mizuno, T. Transcription of ST2A encoding a sulfotransferase family protein that is involved in jasmonic acid metabolism is controlled according to the circadian clock- and PIF4/PIF5-mediated external coincidence mechanism in Arabidopsis thaliana. Biosci. Biotech. Bioch. 77, 2454–2460 (2013).
Oh, E., Zhu, J.-Y. & Wang, Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802 (2012).
Leivar, P. & Quail, P. H. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28 (2011).
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C. & Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323 (2008).
Li, L. et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790 (2012).
Hirschmann, F., Krause, F. & Papenbrock, J. The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Front. Plant Sci. 5, 556 (2014).
Koprivova, A. & Kopriva, S. Sulfation pathways in plants. Chem. Biol. Int. 259, 23–30 (2016).
Gidda, S. K. et al. Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278, 17895–17900 (2003).
Baumann, E. Ueber aulfosäuren im harn. Ber. Dtsch. Chem. Ges. 9, 54–58 (1876).
Gamage, N. et al. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 90, 5–22 (2006).
Komori, R., Amano, Y., Ogawa-Ohnishi, M. & Matsubayashi, Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl Acad. Sci. USA 106, 15067–15072 (2009).
Agrawal, A., Kearney, E., Hastings, A. & Ramsey, T. Attenuation of the jasmonate burst, plant defensive traits, and resistance to specialist monarch caterpillars on shaded common milkweed (Asclepias syriaca). J. Chem. Ecol. 38, 893–901 (2012).
Acosta, I. F. et al. Role of NINJA in root jasmonate signaling. Proc. Natl Acad. Sci. USA 110, 15473–15478 (2013).
Heitz, T., Smirnova, E., Marquis, V. & Poirier, L. Metabolic control within the jasmonate biochemical pathway. Plant Cell Physiol. 60, 2621–2628 (2019).
Smirnova, E. et al. Jasmonic acid oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea infection. Mol. Plant 10, 1159–1173 (2017).
Halkier, B. A. & Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 (2006).
Mugford, S. G. et al. Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21, 910–927 (2009).
Yan, Y. et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483 (2007).
Zhang, Y. & Turner, J. G. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3, e3699 (2008).
Robson, F. et al. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22, 1143–1160 (2010).
de Wit, M. et al. Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 75, 90–103 (2013).
de Wit, M., Ljung, K. & Fankhauser, C. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 208, 198–209 (2015).
Park, J.-H. et al. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12 (2002).
Fernández-Calvo, P. et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715 (2011).
Zhang, T. et al. Hormone crosstalk in wound stress response: wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 67, 2107–2120 (2015).
Schumacher, P. et al. A phosphorylation switch turns a positive regulator of phototropism into an inhibitor of the process. Nat. Commun. 9, 2403 (2018).
Zhang, B. et al. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 22, 26759 (2017).
Ballaré, C. L., Scopel, A. L. & Sánchez, R. A. Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science 247, 329–332 (1990).
Mazza, C. A. & Ballaré, C. L. Photoreceptors UVR8 and phytochrome B cooperate to optimize plant growth and defense in patchy canopies. New Phytol. 207, 4–9 (2015).
Cerrudo, I. et al. Exploring growth-defence trade-offs in Arabidopsis: phytochrome B inactivation requires JAZ10 to suppress plant immunity but not to trigger shade-avoidance responses. Plant Cell Environ. 40, 635–644 (2017).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001).
Li, C. & Wong, W. H. in The Analysis of Gene Expression Data: Methods and Software (eds. Parmigiani, G. et al.) 120–141 (Springer, 2003).
Gendrel, A. V., Lippman, Z., Martienssen, R. & Colot, V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218 (2005).
Hornitschek, P., Lorrain, S., Zoete, V., Michielin, O. & Fankhauser, C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902 (2009).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551 (2013).
Vadassery, J. et al. CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175 (2012).
Koo, A. J. et al. Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-l-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J. Biol. Chem. 289, 29728–29738 (2014).
Koo, A. J. K., Cooke, T. F. & Howe, G. A. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc. Natl Acad. Sci. USA 108, 9298–9303 (2011).
Heitz, T. et al. in Lipids in Plant and Algae Development (eds. Nakamura, Y. & Li-Beisson, Y.) 405–426 (Springer, 2016).
Burow, M., Muller, R., Gershenzon, J. & Wittstock, U. Altered glucosinolate hydrolysis in genetically engineered Arabidopsis thaliana and its influence on the larval development of Spodoptera littoralis. J. Chem. Ecol. 32, 2333–2349 (2006).
Cargnel, M. D., Demkura, P. V. & Ballaré, C. L. Linking phytochrome to plant immunity: low red:far-red ratios increase Arabidopsis susceptibility to Botrytis cinerea by reducing the biosynthesis of indolic glucosinolates and camalexin. New Phytol. 204, 342–354 (2014).
Jeschke, V., Gershenzon, J. & Vassão, D. G. A mode of action of glucosinolate-derived isothiocyanates: detoxification depletes glutathione and cysteine levels with ramifications on protein metabolism in Spodoptera littoralis. Insect Biochem. Mol. Biol. 71, 37–48 (2016).
Piotrowski, M. et al. Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279, 50717–50725 (2004).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Acknowledgements
We thank I. Cerrudo, P. Karssemeijer and B. F. Alliani for help with the initial characterization of st2a lines and P. V. Demkura, E. Goschala, B. Rothe, D. Veit and A. Weber for excellent technical assistance. This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica, Universidad de Buenos Aires and The New Phytologist Trust (C.L.B. and A.T.A.), the Max Planck Society (J.G.), National Science Foundation (grant no. IOS-1557439, A.J.K.), University of Lausanne and Swiss National Science Foundation (C.F.), a Georg Forster Research Award from the Alexander von Humboldt Foundation (C.L.B.) and a Deutscher Akademischer Austauschdienst Fellowship (G.L.F.-M.).
Author information
Authors and Affiliations
Contributions
G.L.F.-M. contributed to all aspects of this research. C.D.C. contributed to experimental design, data collection, analysis and interpretation. M.R. designed and executed protocols for metabolite and hormone analyses. T.Z. and A.J.K. designed and performed hormone profiling. M.D.C. carried out bioassays. M.Z.L. screened mutant lines and helped with gene expression analyses. C.A.M. and T.G.K. performed analyses of transcriptomic data and helped with data interpretation. A.-S.F. and C.F. performed ChIp–qrtPCR experiments. A.T.A. and J.G. contributed to the general conception of the project and data interpretation. C.L.B. conceived the project and contributed to data generation and analysis, and wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
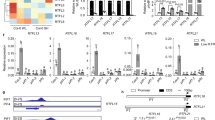
Extended Data Fig. 1 AT5G07010 (ST2a) belongs to a small cluster of MeJA-upregulated genes that are also upregulated under conditions of phyB inactivation.
Heat map (upper part) showing a cluster of genes (approximately 25 % of the genes upregulated by 200 µM MeJA) that are expressed more strongly in the phyB-9 mutant than in Col-0 plants, and mean expression values + /− 1 SD (lower part) of this set of genes. This cluster contains AT5G07010 (ST2a) (for a full list of genes in this cluster, see Supplementary Data File 3). b, Examination of publicly available microarray data shows that among the genes included in categories with GO terms that contain the word “jasmonic”, AT5G07010 stands out for being the most strongly upregulated gene in plants exposed to low R:FR ratios (see arrow). For each gene, dots indicate the average of seven independent microarray experiments (see Methods for details); bars indicate 95 % confidence intervals. c, qPCR analysis demonstrated strong upregulation of ST2a in response to supplemental FR radiation and downregulation by supplemental UVB radiation under our experimental conditions (5-week-old Col-0 plants; relative expression data were normalized to the mean of the Ambient control). Bars indicate treatment means; thin bars = 1 SE; n = 4 biological replicates (small open circles); differences between light treatments tested by one-way ANOVA.
Extended Data Fig. 2 Upregulation of ST2a expression is mediated by PIFs and MYCs in response to low R:FR ratio and wounding, respectively.
a, In a pif4pif5pif7 triple mutant, which does not activate morphological responses to low R:FR ratios, the transcription of ST2a was normally upregulated by mechanical wounding, but not at all by supplemental FR radiation. GxL indicates the significance of the genotype x light interaction term in wounded plants (two-way ANOVA), and different letters indicate significant differences between means (Tukey). b, Effect of FR supplementation on ST2a expression in multiple pif mutants. Different letters indicate significant differences between genotype means in plants exposed to FR. c, Effect of mechanical wounding on ST2a expression in multiple myc mutants. In the wounding treatments, rosettes were harvested 4 h after being mechanically damaged (see Methods). Genotype effects in b and c were tested using one-way ANOVA; different letters indicate significant differences between genotype means (Tukey test). Bars indicate means; thin bars = 1 SE; n = 4 (a) or 3 (b, c) biological replicates (small open circles).
Extended Data Fig. 3 FR regulates the transcription of several genes involved in jasmonate metabolism.
Expression of genes encoding for the following enzymes were measured by qPCR in Col-0 Arabidopsis rosettes exposed to two light conditions (Amb or FR) at different times (0, 0.5, 1, 3 and 6 h) after MeJA (200 μM) treatment: ILL6 and IAR3; JOX/JAO; CYP94B3 and CYP94C1; ST2a; and JAR1. The bar charts show the relative expression data, only for those genes and time points in which FR had a significant (P < 0.05) effect (one-way ANOVA). Bars indicate means; thin bars = 1 SE; at each time point, n = 3 independent biological replicates (small open circles).
Extended Data Fig. 4 FR radiation reduced the concentrations of OH-JA and the sum of JA-Ile conjugates in wounded plants and the concentration of these compounds was higher in st2a-1 than in Col-0.
Bars indicate means; thin bars = 1 SE; at each time point, the interactive effects of genotype and light were tested using two-way ANOVA; n = 5 independent biological replicates (small open circles). Significant (P < 0.05) terms in the factorial analysis are indicated for each panel.
Extended Data Fig. 5 The accumulation of HSO4-JA is determined by the expression of ST2a.
FR fails to upregulate the pool of HSO4-JA in a pif4 pif5 pif7 triple knock-out mutant, where FR fails to upregulate ST2a expression. Samples for metabolite analysis were taken 4 h after wounding. The dotted/dashed lines parallel to the abscissa indicate the basal levels of HSO4-JA in plant of both genotypes harvested before the wounding treatment. Bars indicate means for wounded plants; thin bars = 1 SE; n = 6 independent pools of 3 rosettes each (small open circles). The interactive effect of genotype and light were tested using two-way ANOVA; different letters indicate significant differences between means. b, In Col-0 plants, there is a tight, significant positive correlation between ST2a mRNA and HSO4-JA concentration in rosette tissues (two-tailed Spearman test). Different levels of ST2a gene expression were obtained by varying the amount of mechanical damage (see Methods for details). Samples were taken 4 h after wounding (each datum point represents an independent pool of 3 rosettes).
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, Tables 1 and 2, and methods.
Supplementary Dataset 1
Over-represented GO categories in RNA-seq data reported in Fig. 3b.
Supplementary Dataset 2
Identification codes for sulfotransferase sequences.
Supplementary Dataset 3
List of genes included in the heat map of Extended Data Fig. 1a.
Rights and permissions
About this article
Cite this article
Fernández-Milmanda, G.L., Crocco, C.D., Reichelt, M. et al. A light-dependent molecular link between competition cues and defence responses in plants. Nat. Plants 6, 223–230 (2020). https://doi.org/10.1038/s41477-020-0604-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0604-8
This article is cited by
-
AtBBX29 integrates photomorphogenesis and defense responses in Arabidopsis
Photochemical & Photobiological Sciences (2023)
-
Plant susceptibility to a shared herbivore is reduced by belowground competition with neighbors
Oecologia (2023)
-
PIFs- and COP1-HY5-mediated temperature signaling in higher plants
Stress Biology (2022)
-
Sulfated phenolic acids in plants
Planta (2022)
-
Warm temperature triggers JOX and ST2A-mediated jasmonate catabolism to promote plant growth
Nature Communications (2021)