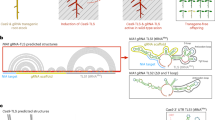
Abstract
Long-distance RNA movement is important for plant growth and environmental responses; however, the extent to which RNAs move between distant tissues, their relative magnitude and functional importance remain to be elucidated on a genomic scale. Using a soybean (Glycine max)–common bean (Phaseolus vulgaris) grafting system, we identified 100 shoot–root mobile microRNAs and 32 shoot–root mobile phased secondary small interfering RNAs (phasiRNAs), which were predominantly produced in shoots and transported to roots, and, in most cases, accumulated to a level similar to that observed in shoots. Many of these microRNAs or phasiRNAs enabled cleavage of their messenger RNA targets or phasiRNA precursors in roots. In contrast, most mobile-capable mRNAs were transcribed in both shoots and roots, with only small proportions transported to recipient tissues. These findings suggest that the regulatory mechanisms for small RNA movement are different from those for mRNA movement, and that the former is more strictly regulated and, probably, more functionally important than the latter.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The raw read sequences are deposited in the National Center for Biotechnology Information Sequence Read Achieve (http://www.ncbi.nlm.nih.gov/sra/) under the accession number PRJNA648759. The miRNAbase53 database (Release 22.1, 2018) was used for identifying known soybean and common bean miRNAs and their precursors.
Code availability
Custom Perl codes used for data analysis in this paper can be found at https://github.com/wang3283/sRNA-normalize-and-GO-anlaysis-input.
References
Wang, J., Jiang, L. & Wu, R. Plant grafting: how genetic exchange promotes vascular reconnection. N. Phytol. 214, 56–65 (2017).
Kehr, J. & Kragler, F. Long distance RNA movement. N. Phytol. 218, 29–40 (2018).
Liu, L. & Chen, X. Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4, 869–878 (2018).
Lee, J. Y. Plasmodesmata: a signaling hub at the cellular boundary. Curr. Opin. Plant Biol. 27, 133–140 (2015).
Aloni, B., Cohen, R., Karni, L., Aktas, H. & Edelstein, M. Hormonal signaling in rootstock–scion interactions. Sci. Hortic. 127, 119–126 (2010).
Zhang, S., Sun, L. & Kragler, F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150, 378–387 (2009).
Thieme, C. J. et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1, 15025 (2015).
Ruiz-Medrano, R., Xoconostle-Cázares, B. & Lucas, W. J. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126, 4405–4419 (1999).
Banerjee, A. K. et al. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457 (2006).
Lu, K. J., Huang, N. C., Liu, Y. S., Lu, C. A. & Yu, T. S. Long-distance movement of Arabidopsis FLOWERING LOCUS T RNA participates in systemic floral regulation. RNA Biol. 9, 653–662 (2012).
Mahajan, A., Bhogale, S., Kang, I. H., Hannapel, D. J. & Banerjee, A. K. The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant Mol. Biol. 79, 595–608 (2012).
Notaguchi, M., Wolf, S. & Lucas, W. J. Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 54, 760–772 (2012).
Zhang, C. et al. Rhizobial infection triggers systemic transport of endogenous RNAs between shoots and roots in soybean. Sci. China Life Sci. 63, 1213–1226 (2020).
Yoo, B. C. et al. A systemic small RNA signaling system in plants. Plant Cell 16, 1979 (2004).
Buhtz, A., Springer, F., Chappell, L., Baulcombe, D. C. & Kehr, J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53, 739–749 (2008).
Pant, B. D. et al. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541–1555 (2009).
Ham, B. K. & Lucas, W. J. Phloem-mobile RNAs as systemic signaling agents. Annu Rev. Plant Biol. 68, 173–195 (2017).
Lin, S. I. et al. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732 (2008).
Pant, B. D., Buhtz, A., Kehr, J. & Scheible, W. R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738 (2008).
Buhtz, A., Pieritz, J., Springer, F. & Kehr, J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10, 64 (2010).
Tsikou, D. et al. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233 (2018).
Molnar, A. et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875 (2010).
Lewsey, M. G. et al. Mobile small RNAs regulate genome-wide DNA methylation. Proc. Natl Acad. Sci. USA 113, E801–E810 (2016).
Vazquez, F. et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79 (2004).
Allen, E., Xie, Z., Gustafson, A. M. & Carrington, J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005).
Chitwood, D. H. et al. Pattern formation via small RNA mobility. Genes Dev. 23, 549–554 (2009).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010).
Schmutz, J. et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46, 707–713 (2014).
Melnyk, C. W., Schuster, C., Leyser, O. & Meyerowitz, E. M. A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr. Biol. 25, 1306–1318 (2015).
Zhang, Z., Liu, X., Guo, X., Wang, X. J. & Zhang, X. Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nat. Plants 2, 16049 (2016).
Kurihara, Y. & Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA 101, 12753–12758 (2004).
Zhai, J. et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553 (2011).
Boualem, A. et al. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 54, 876–887 (2008).
Zhao, M., Meyers, B. C., Cai, C., Xu, W. & Ma, J. Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean. Plant Cell 27, 546–562 (2015).
Fei, Q. et al. Biogenesis of a 22-nt microRNA in Phaseoleae species by precursor-programmed uridylation. Proc. Natl Acad. Sci. USA 115, 8037–8042 (2018).
Liang, D., Rosemary, G. W. & Peter, M. W. Gene silencing in Arabidopsis spreads from the root to the shoot, through a gating barrier, by template-dependent, nonvascular, cell-to-cell movement. Plant Physiol. 159, 984–1000 (2012).
Xia, C., Huang, J., Lan, H. & Zhang, C. Long-distance movement of mineral deficiency-responsive mRNAs in Nicotiana benthamiana/tomato heterografts. Plants 9, 876 (2020).
Sessegolo, C. et al. Transcriptome profiling of mouse samples using nanopore sequencing of cDNA and RNA molecules. Sci. Rep. 9, 14908 (2019).
Narsai, R. et al. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19, 3418–3436 (2007).
Xia, C. et al. Elucidation of the mechanisms of long-distance mRNA movement in a Nicotiana benthamiana/tomato heterograft system. Plant Physiol. 177, 745 (2018).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The sequence alignment/map format and SAM tools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009).
Lin, F. et al. Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC Genomics 15, 18 (2014).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Arikit, S. et al. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26, 4584–4601 (2014).
Thody, J. et al. PAREsnip2: a tool for high-throughput prediction of small RNA targets from degradome sequencing data using configurable targeting rules. Nucleic Acids Res. 46, 8730–8739 (2018).
Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 8, 155–162 (2019).
Guo, Q., Qu, X. & Jin, W. PhaseTank: genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics 31, 284–286 (2015).
Dai, X., Zhuang, Z. & Zhao, P. X. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54 (2018).
Ren, B., Wang, X., Duan, J. & Ma, J. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922 (2019).
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F. & Hellens, R. P. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12 (2007).
Libault, M. et al. Identification of four soybean reference genes for gene expression normalization. Plant Genome 1, 44–54 (2008).
Borges, A., Tsai, S. M. & Caldas, D. G. G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 31, 827–838 (2012).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Acknowledgements
This work was partially supported by the Agriculture and Food Research Initiative of the USDA National Institute of Food and Agriculture (grant no. 2018-67013-27425 to J.M.), the National Natural Science Foundation of China (grant no. 31971898 to S.L.), the National Key R&D Project of China (grant no. 2016YFD0100304 to C.C. and S.L.) and the Purdue AgSEED programme to J.M.
Author information
Authors and Affiliations
Contributions
J.M., S.L. and X.W. designed the research. X.W. analysed the data. S.L., X.W., W.X., T.L., C.C., L.C. and C.B.C. performed the research. J.M. wrote the manuscript with input from X.W. and S.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Margaret Frank, Charles Melnyk and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Relative abundances of both the mobile and immobile sRNAs in grafted shoot and root tissues.
a. Comparison of soybean-specific hcsiRNA abundances between the Pv/Gm and Gm/Gm roots. b, Comparison of common bean-specific hcsiRNA abundances between the Gm/Pv and Pv/Pv roots. c. Comparison of soybean-specific miRNA abundances between the Pv/Gm and Gm/Gm roots. d, Comparison of common bean-specific miRNA abundances between the Gm/Pv and Pv/Pv roots. e. Comparison of soybean-specific phasiRNA abundances between the Pv/Gm and Gm/Gm roots. f, Comparison of common bean-specific phasiRNA abundances between the Gm/Pv and Pv/Pv roots. Gm and Pv indicate soybean and common bean, respectively, and the specific tissue in two grafted samples is underlined below using the shoot/root notation.
Extended Data Fig. 2 Relative abundance of mobile hcsiRNAs in grafted shoot and root tissues.
a. Comparison of soybean hcsiRNA abundances between the Gm/Pv shoots and the Gm/Pv roots. b, Comparison of common bean hcsiRNA abundances between the Pv/Gm shoots and the Pv/Gm roots. c, Comparison of soybean hcsiRNA abundances between the Gm/Gm and the Gm/Pv shoots. d, Comparison of common bean hcsiRNA abundances between the Pv/Pv and Pv/Gm shoots. Gm and Pv indicate soybean and common bean, respectively, and the specific tissue in two grafted samples is underlined below using the shoot/root notation.
Extended Data Fig. 3 Relative abundances of a representative subset of mobile miRNAs in grafted shoot and root tissues validated by stem-loop qRT-PCR.
Gm and Pv indicate soybean and common bean, respectively, and two specific tissue in grafted samples is underlined below using the shoot/root notation. Values are shown as means ± standard errors (s.e.) from three biological replicates. All statistically differences between means with p < 0.01 are indicated as different letters on barplots (one-way ONOVA, Tukey HSD).
Extended Data Fig. 4 Relative abundance of mobile miRNAs and MiRNA transcripts in grafted tissues.
a. Comparison of soybean miRNA abundances between the Gm/Pv shoots and the Gm/Pv roots. b, Comparison of common bean miRNA abundances between the Pv/Gm shoots and the Pv/Gm roots. c, Comparison of soybean miRNA abundances between the Gm/Gm sand the Gm/Pv shoots. d, Comparison of common bean miRNA abundances between the Pv/Pv and the Pv/Gm shoots. e, Comparison of soybean MiRNA abundances between the Gm/Pv shoots and the Pv/Gm roots. f, Comparison of common bean MiRNA abundances between the Pv/Gm shoots and the Gm/Pv roots. Gm and Pv indicate soybean and common bean, respectively, and the specific tissue in two grafted samples is underlined below using the shoot/root notation.
Extended Data Fig. 5 Relative abundance and of mobile phasiRNAs and PHAS transcripts in grafted tissues.
a, Comparison of soybean phasiRNA abundances between the Gm/Pv shoots and the Gm/Pv roots. b, Comparison of common bean phasiRNA abundances between the Pv/Gm shoots and the Pv/Gm roots. c, Comparison of soybean phasiRNA abundances between the Gm/Gm and the Gm/Pv shoots. d, Comparison of common bean phasiRNA abundances between the Pv/Pv and the Pv/Gm shoots. e, Comparison of soybean PHAS abundances between the Gm/Pv shoots and the Pv/Gm roots. f, Comparison of common bean PHAS abundances between the Pv/Gm shoots and the Gm/Pv roots. Gm and Pv indicate soybean and common bean, respectively, and the specific tissue in two grafted samples is underlined below using the shoot/root notation.
Extended Data Fig. 6 Relative abundance of a representative subset of mobile mRNAs validated by qRT-PCR.
Gm and Pv indicate soybean and common bean, respectively, and the specific tissue in two grafted samples is underlined below using the shoot/root notation. Values are shown as means ± standard errors (s.e.) from three biological replicates. All statistically differences between means with p < 0.01 are indicated as different letters on barplots (one-way ONOVA, Tukey HSD).
Extended Data Fig. 7 Gene ontology (GO) terms enrichment of mobile mRNAs in soybean and common bean.
“S->R” indicates the shoot-to-root movement of mRNAs, whereas “R->S” indicates the root-to-shoot movement of mRNAs. GO slims, the cut-down versions of the GO ontologies containing a subset of the terms in the whole GO in plants were used for the enrichment analysis.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 3.
Supplementary Tables
Supplementary Tables 2 and 4–13.
Supplementary Data 1
Soybean and common bean mobile sRNAs detected by grafting.
Rights and permissions
About this article
Cite this article
Li, S., Wang, X., Xu, W. et al. Unidirectional movement of small RNAs from shoots to roots in interspecific heterografts. Nat. Plants 7, 50–59 (2021). https://doi.org/10.1038/s41477-020-00829-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-00829-2
This article is cited by
-
Identification and expression analysis of SQUAMOSA promoter-binding protein (SBP) genes in mungbean
Plant Biotechnology Reports (2023)
-
Mechanism and stability of low cadmium accumulation in grafted soybeans induced by rootstocks
Plant and Soil (2023)
-
Genome-wide identification and characterization of mungbean CIRCADIAN CLOCK ASSOCIATED 1 like genes reveals an important role of VrCCA1L26 in flowering time regulation
BMC Genomics (2022)
-
Plant gene silencing signals move from the phloem to influence gene expression in shoot apical meristems
BMC Plant Biology (2022)
-
Plant and animal small RNA communications between cells and organisms
Nature Reviews Molecular Cell Biology (2022)