Abstract
The blue-light receptor cryptochrome (CRY) in plants undergoes oligomerization to transduce blue-light signals after irradiation, but the corresponding molecular mechanism remains poorly understood. Here, we report the cryogenic electron microscopy structure of a blue-light-activated CRY2 tetramer at a resolution of 3.1 Å, which shows how the CRY2 tetramer assembles. Our study provides insights into blue-light-mediated activation of CRY2 and a theoretical basis for developing regulators of CRYs for optogenetic manipulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Atomic coordinates and structure factors of blue light activated CRY2 tetramer have been deposited in the Protein Data Bank under accession codes PDB 6M79. The cryo-EM map of this tetramer has been deposited in the EM Database with accession code EMD-30128. Source data for Fig. 1b, Extended Data Figs. 6, 7 and 9, and Supplementary Fig. 4 are provided with this paper online. The structures of CRY2N monomer under darkness (PDB accession code 6K8I), BIC2-CRY2N complex (PDB accession code 6K8K), and ZmCRY1cW368A tetramer (PDB accession code 6LZ3) used in this study had been reported (https://doi.org/10.1038/s41594-020-0410-z and https://doi.org/10.1038/s41594-020-0420-x). Source data are provided with this paper.
References
Cashmore, A. R. et al. Cryptochromes: blue light receptors for plants and animals. Science 284, 760–765 (1999).
Wang, Q. & Lin, C. T. Mechanisms of cryptochrome-mediated photoresponses in plants. Annu. Rev. Plant Biol. 71, 23.1–23.27 (2020).
Wang, Q. et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354, 343–347 (2016).
Ma, L. et al. Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2. Nat. Struct. Mol. Biol. 27, 472–479 (2020).
Shao, K. et al. The oligomeric structures of plant cryptochromes. Nat. Struct. Mol. Biol. 27, 480–488 (2020).
Liu, Q. et al. Photooligomerization determines photosensitivity and photoreactivity of plant cryptochromes. Mol. Plant 13, 398–413 (2020).
Brautigam, C. A. et al. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 101, 12142–12147 (2004).
Lin, C. T. et al. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–970 (1995).
Liu, B. B. et al. Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J. Plant Res. 129, 137–148 (2016).
Banerjee, R. et al. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem. 282, 14916–14922 (2007).
Bouly, J. P. et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282, 9383–9391 (2007).
Liu, H. T. et al. The action mechanisms of plant cryptochromes. Trends Plant Sci. 16, 684–691 (2011).
Wang, X. et al. Cryptochrome-mediated light responses in plants. Enzymes 35, 167–189 (2014).
Hense et al. Proton transfer to flavin stabilizes the signaling state of the blue light receptor plant cryptochrome. J. Biol. Chem. 290, 1743–1751 (2015).
Ahmad, M. & Cashmore, A. R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166 (1993).
Guo, H. W. et al. Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363 (1998).
El-Din El-Assal, S. et al. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29, 435–440 (2001).
Hallett, R. A. et al. Correlating in vitro and in vivo activities of light-inducible dimers: a cellular optogenetics guide. ACS Synth. Biol. 5, 53–64 (2016).
Yu, X. H. et al. Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21, 118–130 (2009).
Zuo, Z. C. et al. A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol. Plant 5, 726–733 (2012).
Yang, L. et al. Using HEK293T expression system to study photoactive plant cryptochromes. Front. Plant Sci. 7, 940–947 (2016).
Ozkan-Dagliyan, I. et al. Formation of Arabidopsis cryptochrome 2 photobodies in mammalian nuclei. J. Biol. Chem. 288, 23244–23251 (2013).
Durrant, J. D. et al. POVME 2.0: an enhanced tool for determining pocket shape and volume characteristics. J. Chem. Theory Comput. 10, 5047–5056 (2014).
Ganguly, A. et al. Changes in active site histidine hydrogen bonding trigger cryptochrome activation. Proc. Natl Acad. Sci. USA 113, 10073–10079 (2016).
Berntsson, O. et al. Photoactivation of Drosophila melanogaster cryptochrome through sequential conformational transitions. Sci. Adv. 5, eaaw1531 (2019).
Sang, Y. et al. N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17, 1569–1584 (2005).
Ahmad, M. et al. Photocycle and signaling mechanisms of plant cryptochromes. Curr. Opin. Plant Biol. 33, 108–115 (2016).
Li, X. et al. Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (Trp) triad-dependent photoreduction. Proc. Natl Acad. Sci. USA 108, 20844–20849 (2011).
Taslimi, A. et al. Optimized second-generation CRY2–CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 12, 425–430 (2016).
Taslimi, A. et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925 (2014).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171.e14 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Procopio et al. Kinetic modeling of the Arabidopsis cryptochrome photocycle: FADH(o) accumulation correlates with biological activity. Front. Plant Sci. 7, 888–904 (2016).
Langenbacher et al. Microsecond light-induced proton transfer to flavin in the blue light sensor plant cryptochrome. J. Am. Chem. Soc. 131, 14274–14280 (2009).
Acknowledgements
We are grateful to the Cryo-EM Facility Center of Southern University of Science and Technology for providing technical support during EM image acquisition. We thank the research associates at the Center for Protein Research and Public Laboratory of Electron Microscopy, Huazhong Agricultural University, for technical support. We also thank X. L. Qu, College of Horticulture and Forestry Sciences of Huazhong Agricultural University, for technical support on fluorescence microscopy. We thank Q. Wang from Fujian Agriculture and Forestry University for her constructive suggestions during paper preparation. This work was supported by funds from the Ministry of Science and Technology of China (2018YFA0507700), the National Natural Science Foundation of China (31722017 and 31870753), the Fok Ying-Tong Education Foundation (151021), and the Fundamental Research Funds for the Central Universities (2662017PY031).
Author information
Authors and Affiliations
Contributions
P.Y. conceived the project. L.M. and P.Y. designed all the experiments. L.M. performed all the experiments. Q.W., L.M. and J.W. prepared the Cryo-EM grids. X.G. and X.Y. collected the EM data. Z.G. determined the structure. Z.W. calculated the cavity volume. All authors analysed the data and contributed to manuscript preparation. L.M. and P.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM data processing of CRY2 tetramer.
Processing of 3036 movie stacks resulted in a total number of 3,252,248 particles. After 2D classification, 3,178,552 particles were kept and subjected to 3D classifications. The best 209,604 particles from one class were retained for further 3D auto-refinement for which C2 symmetry was applied, resulting in a 3.1 Å density map estimated on the basis of the gold-standard Fourier shell correlation with 0.143 criterion.
Extended Data Fig. 2 Cryo-EM map of CRY2 tetramer generation and resolution estimation.
a, Representative Cryo-EM micrograph of CRY2 oligomer. Experiments were independently repeated three times with similar results. b, Selected reference-free 2D classification averages. 2D classification of CRY2 oligomer particles representing tetramer-like fold. c, Angular distribution plot for the final reconstruction from two different views. d, Local resolution of the Cryo-EM map generated by ResMap colored from blue to red to indicate resolution from high to low (side bar). e, Gold-standard Fourier Shell Correlation curve for CRY2 tetramer reconstruction. f, Fourier Shell Correlation curve of the model versus the map used for model refinement.
Extended Data Fig. 3 Representative Cryo-EM map for key components of the CRY2N tetramer.
a, Cryo-EM density for reduced FAD and interacting residues. b, Cryo-EM density for AMP and interacting residues. c, Cryo-EM density for CRY2N tetramer helices and sheets.
Extended Data Fig. 4 SLS analyses of CRY2 PHR molecular weight in darkness and under blue light.
Static light scattering analyses of CRY2 PHR molecular weight in darkness and under blue light, respectively. All runs were performed in a Superdex 200TM increase 10/300 GL column. The corresponding CRY2 PHR molecular weights for the two conditions were 44±0.24 kDa and 173±1.95 kDa, respectively.
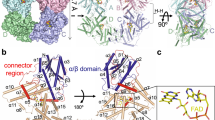
Extended Data Fig. 5 Two interfaces and schematic representation of interactions in the tetramer.
a, Two interfaces of tetramer. CRY2 tetramer is comprised of mol A, B, C, and D that are shown as light pink, cyan, light blue, and yellow, respectively. Connector regions are colored in magenta, red, blue, and green, respectively. Red and black squares show close-up views of interface 1 and interface 2 with 90° rotations, respectively. Numbered helices are involved in the tetramerization. b, Schematic representation of CRY2 tetramer interactions. Extensive interaction networks of interface 1 and interface 2. Residues in mol A, mol B, and mol C involved in interactions are indicated by purple, dark green, and light blue rectangles, respectively. In interface 1, E18 in helix α1; H62 and S66 in helix α3; D160 in helix α6; M167 and R173 in the loop of the connector region; N222 in helix α8; and M267 and R274 in helix α11 from mol A coordinate with M267, R274, R173, M167, D160, N222, E18, H62, and S66 from mol B via hydrogen bonds, respectively (left panel black dashed lines). I17, L159, I163, W172, W214, F262, M267, I270, and I271 interact with each other via hydrophobic interactions (left panel black solid line). Interface 2 is mainly mediated by a dozen hydrogen bonds formed by R50, S202, K329, Q333, R346, W349, E436, R439, A458, and E462 from the two protomers.
Extended Data Fig. 6 Oligomerization state of WT and mutated CRY2 in darkness and under blue light.
a, Oligomerization state of wild type (WT) and mutated CRY2 measured by pull-down assays. Mammalian cells (Expi293F) were co-transfected with constructs designed to express the indicated proteins. The supernatant from lysed cells was incubated with StrepII beads under blue light (50 μmol m−2s−1) or in darkness for 1.5 h. Proteins in input and eluted fractions were detected by immunoblots probed with antibodies against StrepII and Myc. Residues critical for oligomerization are labeled in red. b, Quantification of results in a. The integrated densities of different bands were measured using Image J software. The pull-down prey bands under blue light were normalized by both input and bait bound. Mutants with pull-down bands lower than 20% percent of wild type were shown in red column. The standard deviation is derived from three measurements. Data were presented as mean ± SD. Black circles indicate individual data points for n=3 biological replicates. Differences between each mutants and wild type CRY2 were analyzed by two-tailed paired t-tests. Means with p < 0.05 are indicated. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. Experiments in a were repeated three times, with similar results. Uncropped blot images are available as source data.
Extended Data Fig. 7 Capacity of photobodies formation of different CRY2 mutants.
a, Photobodies formation of CRY2 WT and CRY2 mutants. It was reported that CRY2-GFP, but not GFP-CRY2, forms photobodies in response to blue light19. GFP was constructed to the C-terminal of these CRY2 mutants. B: blue. Bars = 5 μm. b, Percent of cells with photobodies which is calculated by [total number of cells with photobodies in a field of view] / [the number of total cells in the same field view]. The standard deviation is derived from n=80 cells examined 3 independent experiments. Data were presented as mean ± SD. Black circles indicate individual data points for 3 biological replicates. Differences between each mutants and wild type CRY2-GFP were analyzed by two-tailed paired t-tests. Means with p < 0.05 are indicated. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Extended Data Fig. 8 FAD/Reduced FAD cavity volume calculated using POVME.
The FAD/Reduced FAD binding cavities were defined as forming by the residues that possess distances within 4.5 Å to the FAD/Reduced FAD cofactor, and the cavity volume was calculated using POVME23.
Extended Data Fig. 9 SEC analyses of different CRY2 mutants.
All the rans were performed in a SuperoseTM 6 increase 10/300 GL column. Left panel shows SEC analyses of CRY2WT (in darkness and under blue light), CRY2Y232F, CRY2Y232A, CRY2W353A and CRY2W353F. In darkness, wild type CRY2 eluted at 16.5 ml (deep teal line). Whereas, after blue light treatment, activated CRY2 eluted at 14.3 ml (dark purple line), indicating that CRY2 formed a tetramer. In darkness, CRY2Y232A (orange line) and a part of CRY2W353A (peak 1 of red line) eluted at around 14.8 ml, which is similar to that of CRY2WT under blue light, indicating CRY2Y232A and CRY2W353A are tetramers. CRY2Y232F (brown line) and CRY2W353F (green line) were eluted at 16.5 ml, indicating they were monomer. Right panel shows SEC analyses of CRY2WT (in darkness and under blue light), CRY2T244A, CRY2S245A, CRY2N356A and CRY2D387A. Coomassie blue-stained SDS-PAGE gels of peak fractions are on the right of corresponding SEC lines. Experiments were independently repeated three times with similar results. Uncropped gel images are available as source data.
Extended Data Fig. 10 Intra protein hydrophobic interactions of W374 in CRY2N monomer and superposition of structures of blue-light activated CRY2WT tetramer with ZmCRY1W368A tetramer and BIC2-CRY2N complex.
a, CRY2N is shown in white and FAD binding cavity helices are shown in cyan. The black square shows a close-up view of residues that form intra protein hydrophobic interactions with W374. b, Superposition of structures of blue light induced AtCRY2NWT tetramer (in this study) and PHR domain of ZmCRY1W368A tetramer (PDB 6LZ3). c, Structure alignment of CRY2N tetramer (in this study) with BIC2-CRY2N complex (PDB 6K8K). Mol A and D are shown in white; Mol B and C are shown in wheat. BIC2 is shown in red. Surface and ribbon representation of BIC2 is shown. Black dashed lines indicate the clash between BIC2 and interface 2. The black square shows a close-up view of the clash with a 90° rotation.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4, and Tables 1 and 2.
Supplementary Data 1
Unprocessed gels for Supplementary Fig. 4.
Source data
Source Data Fig. 1
Unprocessed gels.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed gels.
Rights and permissions
About this article
Cite this article
Ma, L., Guan, Z., Wang, Q. et al. Structural insights into the photoactivation of Arabidopsis CRY2. Nat. Plants 6, 1432–1438 (2020). https://doi.org/10.1038/s41477-020-00800-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-00800-1
This article is cited by
-
Photoexcited cryptochromes interact with ADA2b and SMC5 to promote the repair of DNA double-strand breaks in Arabidopsis
Nature Plants (2023)
-
Common evolutionary trajectory of short life-cycle in Brassicaceae ruderal weeds
Nature Communications (2023)
-
Direct experimental observation of blue-light-induced conformational change and intermolecular interactions of cryptochrome
Communications Biology (2022)
-
Dynamic physiological and transcriptome changes reveal a potential relationship between the circadian clock and salt stress response in Ulmus pumila
Molecular Genetics and Genomics (2022)
-
The involvement of the N-terminal PHR domain of Arabidopsis cryptochromes in mediating light signaling
aBIOTECH (2021)