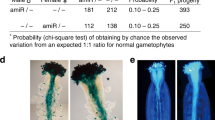
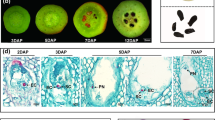
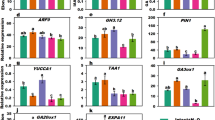
Abstract
The plant hormone ethylene has many roles in growth and development1. In seed plants, the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) is converted into ethylene by ACC oxidase (ACO), and treatment with ACC induces ethylene responses2. However, non-seed plants lack ACO homologues3,4,5,6,7,8, which led us to examine the relationship between ACC and ethylene in the liverwort Marchantia polymorpha. Here, we demonstrate that ACC and ethylene can induce divergent growth responses in Marchantia. Ethylene increases plant and gemma size, induces more gemma cups and promotes gemmae dormancy. As predicted, Mpctr1-knockout mutants display constitutive ethylene responses, whereas Mpein3-knockout mutants exhibit ethylene insensitivity. Compared with the wild type, Mpctr1 gemmae have more and larger epidermal cells, whereas Mpein3 gemmae have fewer and smaller epidermal cells, suggesting that ethylene promotes cell division and growth in developing gemmae. By contrast, ACC treatment inhibits gemma growth and development by suppressing cell division, even in the Mpein3-knockout alleles. Knockout mutants of one or both ACC SYNTHASE (ACS) gene homologues produce negligible levels of ACC, have more and larger gemma cups, and have more-expanded thallus branches. Mpacs2 and Mpacs1 Mpacs2 gemmae also display a high frequency of abnormal apical notches (meristems) that are not observed in ethylene mutants. These findings reveal that ethylene and ACC have distinct functions, and suggest that ACC is a signalling molecule in Marchantia. ACC may be an evolutionarily conserved signal that predates its efficient conversion to ethylene in higher plants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors on reasonable request. Publicly available RNA-seq libraries analysed in this study are listed in Supplementary Data 2. The nucleotide and/or protein sequences analysed in this study are publicly available from NCBI SRA (https://www.ncbi.nlm.nih.gov/sra), NCBI Landmark Database (https://blast.ncbi.nlm.nih.gov/smartblast/smartBlast.cgi?CMD=Web&PAGE_TYPE=BlastDocs#searchSets), Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), GenBank (https://www.ncbi.nlm.nih.gov/genbank/), MarpolBase (https://marchantia.info/), Congenie (http://congenie.org/) and refs. 10,46,47,48,49,50,51,52.
References
Abeles, F. B. M., Morgan, P. W. & Saltveit, M. E. Ethylene in Plant Biology (Academic, 1992).
Yang, S. F. & Hoffman, N. E. Ethylene biosynthesis and its regulation in higher-plants. Ann. Rev. Plant Physiol. 35, 155–189 (1984).
Li, F.-W. et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants 4, 460–472 (2018).
Nishiyama, T. et al. The Chara genome: secondary complexity and implications for plant terrestrialization. Cell 174, 448–464 (2018).
Bowman, J. L. et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304 (2017).
Banks, J. A. et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 (2011).
Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008).
Kawai, Y., Ono, E. & Mizutani, M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 78, 328–343 (2014).
Ju, C. & Chang, C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 169, 85–95 (2015).
Ju, C. et al. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 1, 14004 (2015).
Osborne, D. J., Walters, J., Milborrow, B. V., Norville, A. & Stange, L. M. Evidence for a non-ACC ethylene biosynthesis pathway in lower plants. Phytochemistry 42, 51–60 (1996).
Chernys, J. & Kende, H. Ethylene biosynthesis in Regnellidium diphyllum and Marsilea quadrifolia. Planta 200, 113–118 (1996).
Ishizaki, K., Nishihama, R., Yamato, K. T. & Kohchi, T. Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 57, 262–279 (2016).
Montgomery, S. A. et al. Chromatin organization in early land plants reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Curr. Biol. 30, 573–588 (2020).
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A. & Ecker, J. R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441 (1993).
Chao, Q. et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144 (1997).
Pirrung, M. C., Cao, J. & Chen, J. Ethylene biosynthesis: processing of a substrate analog supports a radical mechanism for the ethylene-forming enzyme. Chem. Biol. 5, 49–57 (1998).
Pirrung, M. C. Ethylene biosynthesis from 1-aminocyclopropanecarboxylic acid. Acc. Chem. Res. 32, 711–718 (1999).
Stange, L. M. D. & Osborne, D. J. In Biochemical and Physiological Aspects of Ethylene Production in Lower and Higher Plants (eds Clijsters, H. et al.) 341–348 (1989).
Kato, H. et al. The roles of the sole activator-type auxin response factor in pattern formation of Marchantia polymorpha. Plant Cell Physiol. 58, 1642–1651 (2017).
Eklund, D. M. et al. Auxin produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. Plant Cell 27, 1650–1669 (2015).
Eklund, D. M. et al. An evolutionarily conserved abscisic acid signaling pathway regulates dormancy in the liverwort Marchantia polymorpha. Curr. Biol. 28, 3691–3699 (2018).
Muday, G. K., Rahman, A. & Binder, B. M. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17, 181–195 (2012).
Uji, T., Endo, H. & Mizuta, H. Sexual reproduction via a 1-aminocyclopropane-1-carboxylic acid-dependent pathway through redox modulation in the marine red alga Pyropia yezoensis (Rhodophyta). Front. Plant Sci. 11, 60 (2020).
Mou, W. et al. Ethylene-independent signaling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat. Commun. 11, 4082 (2020).
Tsang, D. L., Edmond, C., Harrington, J. L. & Nuhse, T. S. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol. 156, 596–604 (2011).
Xu, S. L., Rahman, A., Baskin, T. I. & Kieber, J. J. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20, 3065–3079 (2008).
Vanderstraeten, L., Depaepe, T., Bertrand, S. & Van Der Straeten, D. The ethylene precursor ACC affects early vegetative development independently of ethylene signaling. Front. Plant Sci. 10, 1591 (2019).
Yin, J. et al. Aminocyclopropane-1-carboxylic acid is a key regulator of guard mother cell terminal division in Arabidopsis thaliana. J. Exp. Bot. 70, 897–907 (2019).
Monte, I. et al. An ancient COI-independent function for reactive electrophilic oxylipins in thermotolerance. Curr. Biol. 30, 962–971 (2020).
Fukuda, H., Ogawa, T. & Tanase, S. Ethylene production by micro-organisms. Adv. Microb. Physiol. 35, 275–306 (1993).
John, P. Ethylene biosynthesis: the role of 1-aminocyclopropane-1-carboxylate (ACC) oxidase, and its possible evolutionary origin. Physiol. Plant 100, 583–592 (1997).
Allen, C. J. et al. Cyanobacteria respond to low levels of ethylene. Front. Plant. Sci. 10, 950 (2019).
Yordanova et al. Involvement of ethylene and nitric oxide in cell death in mastoparan-treated unicellular alga Chlamydomonas reinhardtii. Cell Biol. Int. 34, 301–308 (2010).
Vo, T.-T. et al. Effect of the ethylene precursor, 1-aminocyclopropane-1-carboxylic acid on different growth stages of Haematococcus pluvialis. Bioresour. Technol. 220, 85–93 (2016).
Flores-Sandoval, E., Eklund, D. M. & Bowman, J. L. A simple auxin transcriptional response system regulates multiple morphogenetic processes in the liverwort Marchantia polymorpha. PLoS Genet. 11, e1005207 (2015).
NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46, D8–D13 (2018).
Nystedt, B. et al. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584 (2013).
Price, D. C. et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335, 843–847 (2012).
Schonknecht, G. et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210 (2013).
Delwiche, C. F. & Cooper, E. D. The evolutionary origin of a terrestrial flora. Curr. Biol. 25, R899–R910 (2015).
Kakuta, Y. et al. 1-Aminocyclopropane-1-carboxylate synthase of Penicillium citrinum: primary structure and expression in Escherichia coli and Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 65, 1511–1518 (2001).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008).
Vanneste, K., Sterck, L., Myburg, A. A., Van De Peer, Y. & Mizrachi, E. Horsetails are ancient polyploids: evidence from Equisetum giganteum. Plant Cell 27, 1567–1578 (2015).
Zhang, J. et al. The hornwort genome and early land plant evolution. Nat. Plants 6, 107–118 (2020).
Wickett, N. J. et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl Acad. Sci. USA 111, E4859–E4868 (2014).
Hori, K. et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5, 3978 (2014).
Cheng, S. et al. Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179, 1057–1067 (2019).
Wang, S. et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 6, 95–106 (2019).
Cooper, E. & Delwiche, C. Green algal transcriptomes for phylogenetics and comparative genomics. Figshare https://doi.org/10.6084/m9.figshare.1604778 (2016).
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001).
Huelsenbeck, J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. Evolution—Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294, 2310–2314 (2001).
Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005).
Sugano, S. S. et al. Efficient CRISPR-Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS ONE 13, e0205117 (2018).
Ishizaki, K. et al. Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS ONE 10, e0138876 (2015).
Bowman, J. L. et al. The naming of names: guidelines for gene nomenclature in Marchantia. Plant Cell Physiol. 57, 257–261 (2016).
Ishizaki, K., Chiyoda, S., Yamato, K. T. & Kohchi, T. Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091 (2008).
Flores-Sandoval, E., Romani, F. & Bowman, J. L. Co-expression and transcriptome analysis of Marchantia polymorpha transcription factors supports class C ARFs as independent actors of an ancient auxin regulatory module. Front. Plant Sci. 9, 1345 (2018).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids. Res. 46, W537–W544 (2018).
Acknowledgements
We thank Y.-T. Kao for help with maintaining plants; M. Tucker for 1-MCP; and H. Sze for use of her microscope. We acknowledge the Imaging Core Facility in the department of Cell Biology and Molecular Genetics at the University of Maryland, College Park, for the Leica SP5X Laser Scanning Confocal microscope. This work was supported by a NSF grant (no. MCB-1714993, to C.C.), the Australian Research Council (DP170100049, to J.L.B.) and a China Scholarship Council graduate student fellowship (to D.L.). U.A. was supported in part by a grant to the University of Maryland from the Howard Hughes Medical Institute through the Science Education Program. C.C. was supported in part by the Maryland Agricultural Experiment Station.
Author information
Authors and Affiliations
Contributions
All of the authors were involved in aspects of the experimental design. C.C. and J.L.B. conceived and directed the project. D.L. performed almost all of the experiments and produced most of the figures. E.F.-S. generated most of the mutants and conducted in silico analysis of MpACS gene expression. A.C. and U.A. made initial discoveries and performed preliminary experiments. J.M.C. cloned MpACS1 and MpACS2 for yeast expression and generated the ACS tree. J.L.B. generated the CTR1 and EIN3 trees. C.C. wrote the paper with contributions from J.L.B., D.L. and E.F.-S. All of the authors reviewed and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Plants thanks Roberto Salano, Anna Stepanova and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–14, Data 1, Methods 1 and 2, Table 1 and References.
Supplementary Data 2
MpACS1 and MpACS2 expression data from publicly available RNA-seq libraries (source data for Supplementary Fig. 10).
Rights and permissions
About this article
Cite this article
Li, D., Flores-Sandoval, E., Ahtesham, U. et al. Ethylene-independent functions of the ethylene precursor ACC in Marchantia polymorpha. Nat. Plants 6, 1335–1344 (2020). https://doi.org/10.1038/s41477-020-00784-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-00784-y
This article is cited by
-
The effectors of Phytophthora infestans impact host immunity upon regulation of antagonistic hormonal activities
Planta (2023)
-
Genome-wide characterization and identification of candidate ERF genes involved in various abiotic stress responses in sesame (Sesamum indicum L.)
BMC Plant Biology (2022)
-
Phospholipase D activation is required for 1-aminocyclopropane 1-carboxylic acid signaling during sexual reproduction in the marine red alga Neopyropia yezoensis (Rhodophyta)
BMC Plant Biology (2022)
-
The origin of a land flora
Nature Plants (2022)
-
1-Aminocyclopropane-1-carboxylic acid and its analogs alleviate heat stress damage in the marine red alga Neopyropia yezoensis (Rhodophyta)
Journal of Applied Phycology (2022)