Abstract
A key step in microRNA biogenesis is the processing of a primary precursor RNA by the microprocessor into a precursor miRNA (pre-miRNA) intermediate. In plants, little is known about the processes that act on pre-miRNAs to influence miRNA biogenesis. Here, we performed 3′ rapid amplification of complementary DNA ends sequencing to profile pre-miRNA 3′ ends in Arabidopsis. 3′ end heterogeneity was prevalent, and the three microprocessor components promoted 3′ end precision. Extensive cytidylation and uridylation of precise and imprecise pre-miRNA 3′ ends were uncovered. The nucleotidyl transferase HESO1 uridylated pre-miRNAs in vitro and was responsible for most pre-miRNA uridylation in vivo. HESO1, NTP6 and NTP7 contribute to pre-miRNA cytidylation. Tailing of pre-miRNAs tended to restore trimmed pre-miRNAs to their intact length to promote further processing. In addition, HESO1-mediated uridylation led to the degradation of certain imprecisely processed pre-miRNAs. Thus, we uncovered widespread cytidylation and uridylation of pre-miRNAs and demonstrated diverse functions of pre-miRNA tailing in plants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The GEO accession number for the pre-miRNA 3' RACE-seq data is GSE134124. All other data are available in the main text or the Supplementary Information. The data that support the findings of this study are available from the corresponding authors upon request.
References
Xie, M., Zhang, S. & Yu, B. microRNA biogenesis, degradation and activity in plants. Cell. Mol. Life Sci. 72, 87–99 (2015).
Yu, Y., Jia, T. & Chen, X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216, 1002–1017 (2017).
Megha, S., Basu, U. & Kav, N. N. Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 41, 1–15 (2017).
Cui, J., You, C. & Chen, X. The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 35, 61–67 (2017).
Rogers, K. & Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383–2399 (2013).
Yu, B. et al. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935 (2005).
Mi, S. et al. Sorting of small RNAs into Arabidopsis ARGONAUTE complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008).
Baumberger, N. & Baulcombe, D. C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl Acad. Sci. USA 102, 11928–11933 (2005).
Fang, X., Cui, Y., Li, Y. & Qi, Y. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 1, 15075 (2015).
Li, S. et al. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell 30, 481–494 (2018).
Jia, T. et al. The Arabidopsis MOS4-associated complex promotes MicroRNA biogenesis and precursor messenger RNA splicing. Plant Cell 29, 2626–2643 (2017).
Zhang, S., Liu, Y. & Yu, B. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 10, e1004841 (2014).
Zhang, S., Xie, M., Ren, G. & Yu, B. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl Acad. Sci. USA 110, 17588–17593 (2013).
Ren, G. et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 12817–12821 (2012).
Moro, B. et al. Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res. 46, 10709–10723 (2018).
Bologna, N. G. et al. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 23, 1675–1689 (2013).
Dong, Z., Han, M. H. & Fedoroff, N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl Acad. Sci. USA 105, 9970–9975 (2008).
Ré, D. A. et al. Alternative use of miRNA-biogenesis co-factors in plants at low temperatures. Development 146, dev172932 (2019).
Song, J., Song, J., Mo, B. & Chen, x Uridylation and adenylation of RNAs. Sci. China Life Sci. 58, 1057–1066 (2015).
De, A. C., Scheer, H., Zuber, H. & Gagliardi, D. RNA uridylation: a key posttranscriptional modification shaping the coding and noncoding transcriptome. Wiley Interdiscip. Rev. RNA 9, e1440 (2017).
De Almeida, C. et al. RNA uridylation and decay in plants. Phil. Trans. R. Soc. Lond. B 373, 20180163 (2018).
Yang, Z., Ebright, Y. W., Yu, B. & Chen, X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2’ OH of the 3’ terminal nucleotide. Nucleic Acids Res. 34, 667–675 (2006).
Li, J., Yang, Z., Yu, B., Liu, J. & Chen, X. Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507 (2005).
Ren, G., Chen, X. & Yu, B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol. 22, 695–700 (2012).
Zhao, Y. et al. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694 (2012).
Tu, B. et al. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 11, e1005119 (2015).
Wang, X. et al. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3’ tailing of small RNAs in Arabidopsis. PLoS Genet. 11, e1005091 (2015).
Ren, G. et al. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5’ RNA fragments generated by AGO1 cleavage. Proc. Natl Acad. Sci. USA 111, 6365–6370 (2014).
Zuber, H., Scheer, H., Joly, A.-C. & Gagliardi, D. Respective contributions of URT1 and HESO1 to the uridylation of 5′ fragments produced from RISC-cleaved mRNAs. Front. Plant Sci. 9, 1438 (2018).
Heo, I. et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276–284 (2008).
Heo, I. et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708 (2009).
Kim, B. et al. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 34, 1801–1815 (2015).
Chung, W. J. et al. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 21, 286–300 (2011).
Westholm, J. O., Ladewig, E., Okamura, K., Robine, N. & Lai, E. C. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA 18, 177–192 (2012).
Bortolamiol-Becet, D. et al. Selective suppression of the splicing-mediated microRNA pathway by the terminal uridyltransferase Tailor. Mol. Cell 59, 217–228 (2015).
Reimao-Pinto, M. M. et al. Uridylation of RNA hairpins by tailor confines the emergence of MicroRNAs in Drosophila. Mol. Cell 59, 203–216 (2015).
Fang, Y. & Spector, D. L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17, 818–823 (2007).
Zhao, Y., Mo, B. & Chen, X. Mechanisms that impact microRNA stability in plants. RNA Biol. 9, 1218–1223 (2012).
Zhang, H. D., Kolb, F. A., Jaskiewicz, L., Westhof, E. & Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 118, 57–68 (2004).
Zhai, J. et al. Plant microRNAs display differential 3’ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25, 2417–2428 (2013).
Wang, Z. et al. SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557, 516–521 (2018).
Kurihara, Y. & Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA 101, 12753–12758 (2004).
Bologna, N. G., Mateos, J. L., Bresso, E. G. & Palatnik, J. F. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28, 3646–3656 (2009).
Li, S. et al. SMA1, a homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 46, 9148–9159 (2018).
Heo, I. et al. Mono-uridylation of pre-MicroRNA as a key step in the biogenesis of group II let-7 MicroRNAs. Cell 151, 521–532 (2012).
Fei, Q. et al. Biogenesis of a 22-nt microRNA in Phaseoleae species by precursor-programmed uridylation. Proc. Natl Acad. Sci. USA 115, 8037–8042 (2018).
Liu, X. et al. A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis. Mol. Cell 55, 868–879 (2014).
Sement, F. M. et al. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 41, 7115–7127 (2013).
Clarke, J. H., Tack, D., Findlay, K., Van Montagu, M. & Van Lijsebettens, M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 20, 493–501 (1999).
Monaghan, J. et al. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PloS Pathog. 5, e1000526 (2009).
Weihmann, T., Palma, K., Nitta, Y. & Li, X. Pleiotropic regulatory locus 2 exhibits unequal genetic redundancy with its homolog PRL1. Plant Cell Physiol. 53, 1617–1626 (2012).
Wang, X. et al. TCP transcription factors are critical for the coordinated regulation of isochorismate synthase 1 expression in Arabidopsis thaliana. Plant J. 82, 151–162 (2015).
Su, C. et al. The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev. Cell 41, 527–539 e525 (2017).
Popescu, S. C. et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl Acad. Sci. USA 104, 4730–4735 (2007).
Daehwan, K., Ben, L. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Ana, K. & Sam, G. J. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014).
Acknowledgements
We thank Y. Qi for the gift of soe1 and soe2 seeds, B. Zheng for sharing the vector of 35S:HYL1-YFP and Y. Dou for technical assistance. This work was funded by the Guangdong Innovation Team Project (grant no. 2014ZT05S078) and National Science Foundation of China (grant nos. 31571332 and 31560076).
Author information
Authors and Affiliations
Contributions
J.S., X.W., B.M. and X.C. designed the study. J.S., X.W., X.M., L.Y., H.Y. and J.L. performed the experiments. J.S., X.W., B.S., L.G. and X.C. analysed the data. G.R. provided critical reagents and equipment for the study. J.S., X.W., B.M. and X.C. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
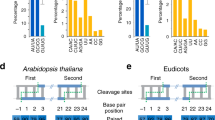
Extended Data Fig. 1 DCL1, HYL1 and SE promote the precise generation of pre-miRNA 3’ ends.
a, Percentage of Class I pre-miRNAs with intact 3’ ends in different genetic backgrounds (replicate 2 and replicate 3). Each dot in the scatter plot represents one pre-miRNA. The reads whose genome-matched 3’ ends were consistent with the annotated ends with or without non-templated addition were defined as ‘intact’. The black and gray horizontal lines represent the median and the quartiles (first and third quartiles), respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Differences between WT (Col-0) and mutants were evaluated by Wilcox. test, two-sided, *P < 0.05; **P < 0.01. See Fig. 1 for biological replicate 1. b, The percentage of individual Class I pre-miRNAs with intact 3’ ends as determined by 3’ RACE-seq in different genetic backgrounds. The reads whose genome-matched 3’ ends were consistent with the annotated ends with or without non-templated addition were defined as ‘intact’. Error bars are s.e.m., n = 3. Differences between WT (Col-0) and mutants were evaluated by student t-tests (paired, two-sided). *P < 0.05; **P < 0.01.
Extended Data Fig. 2 HESO1 confers uridylation and cytidylation of pre-miRNAs.
a–c, HESO1 causes pre-miRNA uridylation and cytidylation. The pHESO1:HESO1 transgene complemented the reduction of pre-miRNA uridylation in heso1-1 (a). Levels of pre-miRNA uridylation (b) and cytidylation (c) in WT (Ler) and heso1-2. Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles, respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage of uridylation or cytidylation was calculated as [number of reads with U or C tails/number of total reads]x100. Differences between WT (Col-0) and heso1 were evaluated by Wilcox. test, two-sided, **P < 0.01. d, Site-specific uridylation analysis of Class I pre-miRNAs in WT (Col-0) and heso1-1. The diagram of positions is shown on the top. Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles, respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage of uridylation was calculated as [number of reads with U tails at a particular position/total number of reads ending at that position]x100. Differences between WT (Col-0) and heso1-1 were evaluated by Wilcox. test, two-sided, **P < 0.01.
Extended Data Fig. 3 Uridylation and cytidylation of pre-miRNAs in Col-0 and heso1-1 (replicate 2 and replicate 3).
a, b, Pre-miRNA uridylation and cytidylation analysis. Levels of pre-miRNA uridylation (a) and cytidylation (b) in WT (Col-0) and heso1-1. c, Cytidylation analysis at 5 positions of Class I pre-miRNAs in WT (Col-0) and heso1-1. Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles (first and third quartiles), respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Differences between WT (Col-0) and heso1-1 were evaluated by Wilcox. test, two-sided, *P < 0.05; **P < 0.01. See Fig. 2 for biological replicate 1.
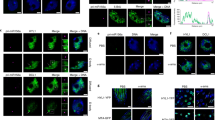
Extended Data Fig. 4 HESO1 localizes in the nucleus.
Sub-cellular localization of HYL1-YFP and HESO1-mCherry in Nicotiana benthamiana epidermal cells. For each construct, 40-50 cells were observed and gave similar patterns of protein localization. YFP and mCherry fluorescence signals were observed by confocal microscopy 40-48 h after agrobacterial infiltration. Scale bar, 20 μm. Experiments were repeated three times and gave similar results.
Extended Data Fig. 5 HESO1 confers site-specific cytidylation of pre-miRNAs.
Error bars are s.e.m., n = 3. Percentage cytidylation was calculated as [number of reads with C tails at a particular position/total number of reads ending at that position]x100. Differences between WT (Col-0) and heso1-1 were evaluated by student t-tests (paired, two-sided). *P < 0.05; **P < 0.01. N.D., not detected.
Extended Data Fig. 6 NTP6 and NTP7 cytidylate pre-miRNAs (replicate 2).
a, Levels of Class I pre-miRNA cytidylation in WT (Col-0) and ten nucleotidyl transferase mutants (replicate 2). Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles (first and third), respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage cytidylation was calculated as [number of reads with C tailing/number of total reads from each pre-miRNA]x100. Differences between WT (Col-0) and mutant were evaluated by Wilcox. test, two-sided, **P < 0.01. b, Analysis of site-specific cytidylation of Class I pre-miRNAs in WT (Col-0), ntp6-1 and ntp7-1(replicate 2). Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles (first and third), respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage cytidylation was calculated as [number of reads with C tailing at a specific 3’ end position/total number of reads with the specific 3’ end position]x100. Differences between WT (Col-0) and mutants were evaluated by Wilcox. test, two-sided, **P < 0.01. c, Histograms showing the degree of cytidylation of individual Class I pre-miRNAs in WT (Col-0), ntp6-1 and ntp7-1. Percentage of cytidylation was calculated as [number of reads with C tails/number of total reads]x100. Error bars are s.e.m., n = 3. Differences between Col-0 and heso1-1 were evaluated by student t-tests (unpaired, two-sided). *P < 0.05. See Fig. 3 for biological replicate 1.
Extended Data Fig. 7 Cytidylation and uridylation of Class I pre-miRNA in nucleotidyl transferase mutants.
a, Phylogenetic analysis of NTP proteins in Arabidopsis. A neighbor-joining (NJ) tree was constructed by MEGA, using the p-distance method with gaps treated by pairwise deletion and a 1,000 bootstrap replicate. b, c, Levels of Class I pre-miRNA cytidylation in WT (Col-0), ntp6 ntp7 and ntp2 ntp6 ntp7. Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles (first and third), respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage cytidylation was calculated as [number of reads with C tailing/number of total reads from each pre-miRNA]x100. Differences between WT (Col-0) and mutant were evaluated by Wilcox. test, two-sided, **P < 0.01. d, Levels of Class I pre-miRNA uridylation in WT (Col-0) and ten nucleotidyl transferase mutants. Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles, respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage of uridylation was calculated as [number of reads with U tailing/number of total reads from each pre-miRNA]x100. Differences between WT (Col-0) and mutant were evaluated by Wilcox. test, two-sided, **P < 0.01.
Extended Data Fig. 8 Quantification of tailing at specific sites for all Class I pre-miRNAs in WT (Col-0) and heso1-1 (replicate 2 and replicate 3).
Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles (first and third), respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage of reads was calculated as [number of reads with specific end type/number of total reads from each pre-miRNA]x100. Differences between WT (Col-0) and heso1-1 were evaluated by Wilcox. test, two-sided, *P < 0.05; **P < 0.01. See Fig. 4 for biological replicate 1.
Extended Data Fig. 9 Tailing status of pre-miRNAs and miRNAs in WT (Col-0) and hen1-8.
a, b, Analysis of uridylation (a) and cytidylation (b) of Class I pre-miRNAs and miRNAs in WT (Col-0) and hen1-8. Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles, respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage of uridylation or cytidylation was calculated to the total reads mapped to each pre-miRNA or miRNA. Mono: mono-uridylation or mono-cytidylation; All: total uridylation or cytidylation. c, d, Analysis of uridylation (c) and cytidylation (d) of individual Class I pre-miRNAs and miRNAs (miRNA locates in the 3 P strand) in WT (Col-0) and hen1-8. Error bars are s.e.m., n = 3. e, f, Percentage of 1-nt and 2-nt tails at position 1 (e) and 2 (f) of pre-miR158a and miR158a in WT (Col-0) and hen1–8. Percentage was calculated based on total tailed pre-miR158a and miR158a at position 1 (e) and 2 (f). The inset shows all cytidine-containing tails. Error bars are s.e.m., n = 3. Differences between WT (Col-0) and hen1–8 were evaluated by student t-tests (unpaired, two-sided). *P < 0.05.
Extended Data Fig. 10 Tailing of pre-miRNAs and miRNAs in WT (Ler) and hen1-2.
a, b, Analysis of uridylation (a) and cytidylation (b) of Class I pre-miRNAs and miRNAs in WT (Ler) and hen1-2. Each dot represents a pre-miRNA. The black and gray lines represent the median and the quartiles, respectively. Values are Class I pre-miRNAs from one biological repeat (n = 24). Percentage of uridylation or cytidylation was calculated to the total reads mapped to each pre-miRNA or miRNA. Mono: mono-uridylation or mono-cytidylation; All: total uridylation or cytidylation. c, d, Analysis of uridylation (c) and cytidylation (d) of individual Class I pre-miRNAs and miRNAs (miRNA locates in the 3 P strand) in WT (Ler) and hen1-2. Error bars are s.e.m., n = 3. e, The uridylation/cytidylation ratio in the tails of pre-miRNAs and miRNAs in WT (Ler) and hen1-2. Tails composed of uridine or cytidine only were included in the quantification of uridylation or cytidylation. Percentage of uridylation or cytidylation was calculated to the total reads mapped to each pre-miRNA and miRNA. Error bars are SEM, n = 3. Differences between WT (Ler) and hen1-2 were evaluated by student t-tests (unpaired, two-sided). *P < 0.05; **P < 0.01. f–h, Percentage of 1-nt and 2-nt tails at position 0 (f), 1 (g) and 2 (h) of pre-miR158a and miR158a in WT (Ler) and hen1-2. Percentage was calculated based on total tailed pre-miR158a and miR158a at position 0 (f), 1 (g) and 2 (h). The inset shows all cytidine-containing tails. Error bars are s.e.m., n = 3. Differences between WT (Ler) and hen1-2 were evaluated by student t-tests (unpaired, two-sided). *P < 0.05.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Unprocessed RNA blots
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Unprocessed images
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 9
Statistical Source Data
Source Data Extended Data Fig. 10
Statistical Source Data
Source Data Supplementary Fig. 1
Statistical Source Data
Source Data Supplementary Fig. 2
Statistical Source Data
Source Data Supplementary Fig. 3
Unprocessed gels and RNA blots.
Rights and permissions
About this article
Cite this article
Song, J., Wang, X., Song, B. et al. Prevalent cytidylation and uridylation of precursor miRNAs in Arabidopsis. Nat. Plants 5, 1260–1272 (2019). https://doi.org/10.1038/s41477-019-0562-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0562-1
This article is cited by
-
MiRNA fine tuning for crop improvement: using advance computational models and biotechnological tools
Molecular Biology Reports (2022)
-
MicroRNA biogenesis in plant
Plant Growth Regulation (2021)
-
NTP4 modulates miRNA accumulation via asymmetric modification of miRNA/miRNA* duplex
Science China Life Sciences (2021)
-
Beyond Dicer’s cut
Nature Plants (2019)