Abstract
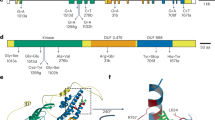
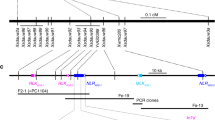
Orobanche cumana (sunflower broomrape) is an obligate parasitic plant that infects sunflower roots, causing yield losses. Here, by using a map-based cloning strategy, we identified HaOr7—a gene that confers resistance to O. cumana race F—which was found to encode a leucine-rich repeat receptor-like kinase. The complete HAOR7 protein is present in resistant lines of sunflower and prevents O. cumana from connecting to the vascular system of sunflower roots, whereas susceptible lines encode a truncated protein that lacks transmembrane and kinase domains.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary Information. GeneBank accession numbers: MN219479, MN219480, MF374791, MF374792.
References
Subrahmaniam, H. J. et al. The genetics underlying natural variation of plant-plant interactions, a beloved but forgotten member of the family of biotic interactions. Plant J. 12, 3218–3221 (2017).
Westwood, J. H., Yoder, J. I., Timko, M. P. & dePamphilis, C. W. The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235 (2010).
Yoshida, S., Cui, S., Ichihashi, Y. & Shirasu, K. The Haustorium, a specialized invasive organ in parasitic plants. Annu. Rev. Plant Biol. 67, 643–667 (2016).
Parker, C. Observations on the current status of orobanche and striga problems worldwide. Pest Manag. Sci. 65, 453–459 (2009).
Li, J. & Timko, M. P. Gene-for-gene resistance in striga-cowpea associations. Science 325, 1094 (2009).
Rispail, N. et al. Plant resistance to parasitic plants: molecular approaches to an old foe. New Phytol. 173, 703–712 (2007).
Louarn, J. et al. Sunflower resistance to Broomrape (Orobanche cumana) is controlled by specific QTLs for different parasitism stages. Front. Plant Sci. 7, 590 (2016).
Pérez-Vich, B. et al. Quantitative trait loci for broomrape (Orobanche cumana Wallr.) resistance in sunflower. Theor. Appl. Genet. 109, 92–102 (2004).
Fernández-Martínez, J. M., Domínguez, J., Pérez-Vich, B. & Velasco, L. Update on breeding for resistance to sunflower broomrape. Helia 33, 73–84 (2008).
Tang, S. et al. Genetic mapping of the gene for resistance to race E in sunflower. Crop Sci. 43, 1021–1028 (2003).
Rodríguez-Ojeda, M. I. et al. A dominant avirulence gene in Orobanche cumana triggers Or5 resistance in sunflower. Weed Res. 53, 322–327 (2013).
Flor, H. H. Current status of the gene-for-gene concept. North 9, 275–296 (1971).
Martín-Sanz, A., Malek, J., Fernández-Martínez, J. M., Pérez-Vich, B. & Velasco, L. Increased virulence in sunflower Broomrape (Orobanche cumana Wallr.) populations from Southern Spain is associated with greater genetic diversity. Front. Plant Sci. 7, 589 (2016).
Molinero-Ruiz, L. et al. History of the race structure of Orobanche cumana and the breeding of sunflower for resistance to this parasitic weed. Span. J. Agric. Res. 13, e10R01 (2015).
Badouin, H. et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152 (2017).
Song, W. et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 2–4 (1995).
Hewitt, E. J. Sand and water culture methods used in the study of plant nutrition (Commonwealth Agricultural Bureau, 1966).
Altschul, S. F., Gish, W., Miller, W., Myers, E. w. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Acknowledgements
We thank all of the following colleagues from LIPM (INRA, France) for their help: E. Sallet for her help in genes annotation, J. Gouzy for his advice on sequence analysis and his reviewing of the manuscript, N. Pouilly for his advice on genotyping experiments, N. Peeters (director of the Toulouse Plant Microbe Phenotyping platform), F. Devoilles and M. Khafif for their help with some phenotyping experiments in the greenhouse. We also thank L. Rieseberg (UBC, Canada) for his reading of the manuscript and for improving the writing. We thank GeT-PlaGe Genomic Platform (INRA-Toulouse, France, https://get.genotoul.fr/en/) for producing PacBio sequence data of the BAC clones. This work was supported by INRA, Syngenta seeds and the French Laboratory of Excellence project TULIP (ANR-10-LABX-41; ANR-11-IDEX-0002-02).
Author information
Authors and Affiliations
Contributions
P.D. contributed to statistical analysis, genotyping, sequencing and phenotyping experiments. S.V. developed BAC libraries and performed their screening. C.C. performed the sequencing of the BAC clones. S.Cauet assembled the sequences of the BAC clones. S.Carrère annotated genes from the genomic sequences. J.B. contributed to phylogenetic analysis. M.-C.B. and F.G. contributed to the crossing and growing of sunflowers. J.-C.R. contributed to the genotyping experiments. M.L.-S. contributed to field phenotyping experiments. M.C. and M.-C.A. performed microscopy experiments. P.P.-E. calculated LD. C.P. contributed to supervising the map-based cloning analysis. B.P.-V. and L.V. contributed to the phenotyping for phylogenetic analysis. H.B. supervised the genomic analysis on BAC clones. J.P. and S.M. designed all experiments and managed the project. All of the authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4, Methods, Data 1–3 and legends for Supplementary Data 1–6.
Supplementary Table 1
Supplementary Tables 1–8.
Supplementary Data 4
Sequences of HanXRQChr07g0202951 on the diversity panel.
Supplementary Data 5
Sequences of HaOr7 (HanXRQChr07g0202981) on the diversity panel.
Supplementary Data 6
Sequences of HanXRQChr07g0202811 on the diversity panel.
Rights and permissions
About this article
Cite this article
Duriez, P., Vautrin, S., Auriac, MC. et al. A receptor-like kinase enhances sunflower resistance to Orobanche cumana. Nat. Plants 5, 1211–1215 (2019). https://doi.org/10.1038/s41477-019-0556-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0556-z
This article is cited by
-
A cluster of putative resistance genes is associated with a dominant resistance to sunflower broomrape
Theoretical and Applied Genetics (2024)
-
Identification and genetic diversity analysis of broomrape in Xinjiang, China
Molecular Biology Reports (2024)
-
Development and characterization of a new sunflower source of resistance to race G of Orobanche cumana Wallr. derived from Helianthus anomalus
Theoretical and Applied Genetics (2024)
-
Phylogeny of the plant receptor-like kinase (RLK) gene family and expression analysis of wheat RLK genes in response to biotic and abiotic stresses
BMC Genomics (2023)
-
HeliantHOME, a public and centralized database of phenotypic sunflower data
Scientific Data (2022)