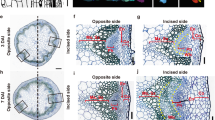
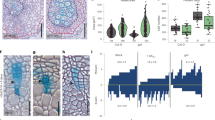
Abstract
Vascular cambium, a lateral plant meristem, is a central producer of woody biomass. Although a few transcription factors have been shown to regulate cambial activity1, the phenotypes of the corresponding loss-of-function mutants are relatively modest, highlighting our limited understanding of the underlying transcriptional regulation. Here, we use cambium cell-specific transcript profiling followed by a combination of transcription factor network and genetic analyses to identify 62 new transcription factor genotypes displaying an array of cambial phenotypes. This approach culminated in virtual loss of cambial activity when both WUSCHEL-RELATED HOMEOBOX 4 (WOX4) and KNOTTED-like from Arabidopsis thaliana 1 (KNAT1; also known as BREVIPEDICELLUS) were mutated, thereby unlocking the genetic redundancy in the regulation of cambium development. We also identified transcription factors with dual functions in cambial cell proliferation and xylem differentiation, including WOX4, SHORT VEGETATIVE PHASE (SVP) and PETAL LOSS (PTL). Using the transcription factor network information, we combined overexpression of the cambial activator WOX4 and removal of the putative inhibitor PTL to engineer Arabidopsis for enhanced radial growth. This line also showed ectopic cambial activity, thus further highlighting the central roles of WOX4 and PTL in cambium development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Gene accession numbers are as follows: ARR15, AT1G74890; CYCD3;1, AT4G34160; ANT, AT4G37750; WOX14, AT1G20700; KNAT1 (also known as BP), AT4G08150; PTL, AT5G03680; SVP, AT2G22540; AGL24, AT4G24540; LBD4, AT1G31320; SCL7, AT3G50650; AGL14, AT4G11880; ATHB53, AT5G66700; ATHB5, AT5G65310; WRI3, AT1G16060; AHL11, AT3G61310; MYB47, AT1G18710; MYB95, AT1G74430; MYB87, AT4G37780; STY2, AT4G36260; RAS1, AT1G09950; AtERF019, AT1G22810; AtERF021, AT1G71450; AtERF029, AT4G25490; AtERF032, AT1G63030; AtERF043, AT4G32800; AtERF071, AT2G47520; AtERF072, AT3G16770; WOX4, AT1G46480; WOX13, AT4G35550; STM, AT1G62360; KNAT2, AT1G70510; KNAT6, AT1G23380; BOP1, AT3G57130; BOP2, AT2G41370; EDA31, AT3G10000; LBD3 (also known as ASL9), AT1G16530; LBD1, AT1G07900; SCL4, AT5G66770; SCL3, AT1G50420; SCL28, AT1G63100; ANAC015 (also known as BRN1), AT1G33280; ANAC070 (also known as BRN2), AT4G10350; SMB, AT1G79580; ANAC042 (also known as JUNGBRUNNEN 1), AT2G43000; ANAC042-like, AT3G12910; MYB3R4, AT5G11510; MYB3R1, AT4G32730; PXY (also known as TDR), AT5G61480; ERECTA/ER, AT2G26330. Raw data of Affymetrix ATH1 GeneChip (accession no. GSE125244) and RNA-seq data can be found in NCBI (accession no. PRJNA523600). Source data related to LithoGraphX analysis (Fig. 3c,d; Supplementary Fig. 11) and ANOVA analysis (Fig. 4d; Supplementary Figs. 8e and 11–13) can be found in Supplementary Datasets. The data that support the findings of this study are available from the corresponding authors upon request. Mutants generated in this study will be deposited to NASC.
Code availability
The code used for network construction and LithoGraphX analysis can be accessed at Github (https://github.com/Zhangcambium2019/Zhang2019). R-codes for boxplot, half-violin plot, median calculation, average calculation and P value calculation can be accessed at Github.
References
Zhang, J., Nieminen, K., Serra, J. A. A. & Helariutta, Y. The formation of wood and its control. Curr. Opin. Plant Biol. 17, 56–63 (2014).
Nieminen, K., Blomster, T., Helariutta, Y. & Mahonen, A. P. Vascular cambium development. Arabidopsis Book 13, e0177 (2015).
Smetana, O. et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565, 485–489 (2019).
Zhang, J., Elo, A. & Helariutta, Y. Arabidopsis as a model for wood formation. Curr. Opin. Biotechnol. 22, 293–299 (2011).
Laux, T., Mayer, K. F. X., Berger, J. & Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96 (1996).
Clark, S. E., Jacobsen, S. E., Levin, J. Z. & Meyerowitz, E. M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122, 1567–1575 (1996).
Benfey, P. N. et al. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119, 57–70 (1993).
Helariutta, Y. et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000).
Lucas, M. et al. SHORT-ROOT regulates primary, lateral and adventitious root development in Arabidopsis. Plant Physiol. 155, 384–398 (2011).
Scheres, B. et al. Mutations affecting the radial organization of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121, 53–62 (1995).
DiLaurenzio, L. et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433 (1996).
Ji, J. B. et al. WOX4 promotes procambial development. Plant Physiol. 152, 1346–1356 (2010).
Hirakawa, Y., Kondo, Y. & Fukuda, H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22, 2618–2629 (2010).
Suer, S., Agusti, J., Sanchez, P., Schwarz, M. & Greb, T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23, 3247–3259 (2011).
Etchells, J. P., Provost, C. M., Mishra, L. & Turner, S. R. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140, 2224–2234 (2013).
Randall, R. S. et al. AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biol. Open 4, 1229–1236 (2015).
Etchells, J. P., Provost, C. M. & Turner, S. R. Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet. 8, e100299 (2012).
Brackmann, K. et al. Spatial specificity of auxin responses coordinates wood formation. Nat. Commun. 9, 875 (2018).
Birnbaum, K. et al. Cell type-specific expression profiting in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619 (2005).
Wagner, G. P., Pavlicev, M. & Cheverud, J. M. The road to modularity. Nat. Rev. Genet. 8, 921–931 (2007).
Wang, X. W., Dalkic, E., Wu, M. & Chan, C. Gene module level analysis: identification to networks and dynamics. Curr. Opin. Biotechnol. 19, 482–491 (2008).
Siligato, R. et al. MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 170, 627–641 (2016).
Uchida, N. & Tasaka, M. Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. J. Exp. Bot. 64, 5335–5343 (2013).
Taylor-Teeples, M. et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–U307 (2015).
Wunderling, A., Ben Targem, M., de Reuille, P. B. & Ragni, L. Novel tools for quantifying secondary growth. J. Exp. Bot. 68, 89–95 (2017).
de Reuille P. B. & Ragni, L. Vascular morphodynamics during secondary growth. Methods Mol. Biol. 1544, 103–125 (2017).
Guo, Y., Qin, G. J., Gu, H. Y. & Qu, L. J. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21, 3518–3534 (2009).
Yordanov, Y. S., Regan, S. & Busov, V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in populus. Plant Cell 22, 3662–3677 (2010).
Etchells, J. P., Mishra, L. S., Kumar, M., Campbell, L. & Turner, S. R. Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050–1055 (2015).
Zhang, J., Serra, J. A. A. & Helariutta, Y. Wood development: growth through knowledge. Nat. Plants 1, 15060 (2015).
Immanen, J. et al. Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr. Biol. 26, 1990–1997 (2016).
Brady, S. M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007).
Nawy, T. et al. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17, 1908–1925 (2005).
Lee, J. Y. et al. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl Acad. Sci. USA 103, 6055–6060 (2006).
Levesque, M. P. et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4, e143 (2006).
Wu, Z. J., Irizarry, R. A., Gentleman, R., Martinez-Murillo, F. & Spencer, F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99, 909–917 (2004).
Smyth, G. K. in Bioinformatics and computational biology solutions using R and Bioconductor (eds Gentleman, R. et al.) 397–420 (Springer, 2005).
Segal, E. et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 34, 166–176 (2003).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
We thank O. Smetana for providing the root cross-section sketches for making vector images in Fig. 1a; S. Miyashima for providing the pBI-nlsYFP-GUS vector for the cloning of pLBD4::nYFP-GUS; H. Fukuda, J. Murray, D. Smyth, U. Fischer, H. Yu, T. Mizuno, J. Lim, B. Scheres, M. Ito, S. Hepworth, V. Pautot, M. Kater and S. Turner for providing published seeds (Supplementary Table 4a,b); K. Kainulainen and M. Herpola for technical assistance; O. Smetana and L.-L. Ye for providing help on finalizing the figures and S. El-Showk for language correction. This work was supported by the Finnish Centre of Excellence in Molecular Biology of Primary Producers (Academy of Finland CoE programme 2014–2019, decision no. 271832), the Gatsby Foundation (no. GAT3395/PR3), the University of Helsinki (award no. 799992091) and the European Research Council Advanced Investigator Grant SYMDEV (no. 323052 to Y.H.); Academy of Finland (grants nos. 132376, 266431 and 271832), University of Helsinki HiLIFE fellowship to A.P.M. and National Research Foundation of Korea (nos. 2018R1A5A1023599 and 2016R1A2B2015258 to J.-Y.L.).
Author information
Authors and Affiliations
Contributions
J.Z., J.-Y.L., A.P.M. and Y.H. designed the experiments. J.Z. carried out most of the experiments with help from other co-authors. K.N. constructed the pARR15::GFP line for FACS. K.N., A.E. and J.-Y.L. performed FACS and microarray hybridization. J.-Y.L. and J.-G.J. analysed the microarray data from the FACS. J.Z. generated and analysed all transgenic lines with input from A.E. on GUS reporter lines. W.Y. and J.Z. carried out the RNA in situ analysis. M.K. conducted the qRT–PCR and analysed the data for network construction. S.E.A. constructed the network and carried out perturbation analysis. J.Z. and G.E. generated combinatorial mutants. J.-Y.Y. and J.-H.L. produced the asl9 CRISPR mutant. J.Z., G.E., J.X. and T.D. performed mutant genotyping. J.Z., with assistance from J.X., G.E. and J.A.-S., characterized the mutant and overexpression line phenotypes. J.Z., G.E. and J.A.-S. conducted quantification and statistical analyses for phenotypic examination of mutants and overexpression lines. J.A.-S. ran the LithoGraphX analysis with input from G.E., L.R. and P.B.R. J.Z. performed the RNA-seq experiment and analysed the RNA-seq data with S.E.A. G.E. drew all box plot figures. J.Z., J.-Y.L., A.P.M. and Y.H. wrote the manuscript with help from the co-authors. All authors read and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
University of Helsinki, University of Cambridge, Natural Resources Institute Finland (Luke) and Seoul National University have filed one patent application (application no. FI0195659) on the use of constructs and methods that are described here to increase radial growth and biomass in plants, in which J.Z., G.E., J. A.-S., M.K., K.N., J.-Y.L., A.P.M. and Y.H. are listed as inventors. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13, Supplementary Tables 1–8, Supplementary Datasets 1–4, Supplementary References and Supplementary Note.
Supplementary Table 1
Cambium enriched gene and gene module identification in Arabidopsis roots.
Supplementary Table 2
Identification of cambium transcription factors.
Supplementary Table 3
Transcript profiling for network and the mutant phenotype prediction.
Supplementary Table 4
Plant materials, cloning vectors and primers used in this study.
Supplementary Table 5
Vascular phenotype characterization in overexpression lines.
Supplementary Table 6
Vascular phenotype characterization of mutants.
Supplementary Table 7
Differentially expressed genes in mutants.
Supplementary Table 8
Cambium genes are perturbed in mutants.
Supplementary Dataset 1
Source data for correlation analyses.
Supplementary Dataset 2
ANOVA output data.
Supplementary Dataset 3
LithoGraphX Cell classifier raw data.
Supplementary Dataset 4
RNA-seq read counts of wild-type (Col) and mutants.
Rights and permissions
About this article
Cite this article
Zhang, J., Eswaran, G., Alonso-Serra, J. et al. Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots. Nat. Plants 5, 1033–1042 (2019). https://doi.org/10.1038/s41477-019-0522-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0522-9
This article is cited by
-
Time course of changes in the transcriptome during russet induction in apple fruit
BMC Plant Biology (2023)
-
Genes expression profiles in vascular cambium of Eucalyptus urophylla × Eucalyptus grandis at different ages
BMC Plant Biology (2023)
-
Trehalose metabolism coordinates transcriptional regulatory control and metabolic requirements to trigger the onset of cassava storage root initiation
Scientific Reports (2023)
-
Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa
Nature Plants (2023)
-
A SHORTROOT-Mediated Transcriptional Regulatory Network for Vascular Development in the Arabidopsis Shoot
Journal of Plant Biology (2022)