Abstract
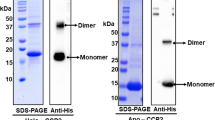
The photoactive orange carotenoid protein (OCP) is a blue-light intensity sensor involved in cyanobacterial photoprotection. Three OCP families co-exist (OCPX, OCP1 and OCP2), having originated from the fusion of ancestral domain genes. Here, we report the characterization of an OCPX and the evolutionary characterization of OCP paralogues focusing on the role of the linker connecting the domains. The addition of the linker with specific amino acids enabled the photocycle of the OCP ancestor. OCPX is the paralogue closest to this ancestor. A second diversification gave rise to OCP1 and OCP2. OCPX and OCP2 present fast deactivation and weak antenna interaction. In OCP1, OCP deactivation became slower and interaction with the antenna became stronger, requiring a further protein to detach OCP from the antenna and accelerate its deactivation. OCP2 lost the tendency to dimerize, unlike OCPX and OCP1, and the role of its linker is slightly different, giving less controlled photoactivation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequences of the OCPs isolated and studied in this article are accessible in the GenBank/EMBL data libraries (or the IMG database) under the following accession numbers: slr1963 (OCP1 Synechocystis, IMG ID: 2514153952), Fdi2450 (OCP1 Tolypothrix, IMG ID: 2501541336), Fdi7374 (OCP2 Tolypothrix, IMG ID: 2501546328) and WA1_RS11680 (OCPX Scytonema, IMG ID: 2551963320). The protein sequences and their corresponding identifiers for the IMG database used for the phylogenetic analysis can be found in the Supplementary Information (available as an .xls file in the online version of the article). The additional data that support the findings of this study are available from the corresponding author upon request.
References
Kirilovsky, D. & Kerfeld, C. A. Cyanobacterial photoprotection by the orange carotenoid protein. Nat. Plants 2, 16180 (2016).
Kerfeld, C. A., Melnicki, M. R., Sutter, M. & Dominguez-Martin, M. A. Structure, function and evolution of the cyanobacterial orange carotenoid protein and its homologs. New Phytol. 215, 937–951 (2017).
Sluchanko, N. N., Slonimskiy, Y. B. & Maksimov, E. G. Features of protein−protein interactions in the cyanobacterial photoprotection mechanism. Biochemistry 82, 1592–1614 (2017).
Kerfeld, C. A. et al. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure 11, 55–65 (2003).
Wilson, A. et al. A photoactive carotenoid protein acting as light intensity sensor. Proc. Natl Acad. Sci. USA 105, 12075–12080 (2008).
Leverenz, R. L. et al. A 12 Å carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection. Science 348, 1463–1466 (2015).
Konold, P. E. et al. Photoactivation mechanism, timing of protein secondary structure dynamics and carotenoid translocation in the orange carotenoid. J. Am. Chem. Soc. 141, 520–530 (2019).
Gupta, S. et al. Local and global structural drivers for the photoactivation of the orange carotenoid protein. Proc. Natl Acad. Sci. USA 112, E5567–E5574 (2015).
Liu, H. et al. Dramatic domain rearrangements of the cyanobacterial orange carotenoid protein upon photoactivation. Biochemistry 55, 1003–1009 (2016).
Maksimov, E. G. et al. The signaling state of orange carotenoid protein. Biophys. J. 109, 595–607 (2015).
Maksimov, E. G. et al. A comparative study of three signaling forms of the orange carotenoid protein. Photosynth. Res. 130, 389–401 (2016).
Wilson, A. et al. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 18, 992–1007 (2006).
Gwizdala, M., Wilson, A. & Kirilovsky, D. In vitro reconstitution of the cyanobacterial photoprotective mechanism mediated by the orange carotenoid protein in Synechocystis PCC 6803. Plant Cell 23, 2631–2643 (2011).
Wilson, C. W. A. et al. The essential role of the N-terminal domain of the orange carotenoid protein in cyanobacterial photoprotection: importance of a positive charge for phycobilisome binding. Plant Cell 24, 1972–1983 (2012).
Boulay, C., Wilson, A., D’haene, S. & Kirilovsky, D. Identification of a protein required for recovery of full antenna capacity in OCP-related photoprotective mechanism in cyanobacteria. Proc. Natl Acad. Sci. USA 107, 11620–11625 (2010).
Sutter, M. et al. Crystal structure of the FRP and identification of the active site for modulation of OCP-mediated photoprotection in cyanobacteria. Proc. Natl Acad. Sci. USA 110, 10022–10027 (2013).
Sluchanko, N. N. et al. The purple Trp288Ala mutant of Synechocystis OCP persistently quenches phycobilisome fluorescence and tightly interacts with FRP. Biochim. Biophys. Acta Bioenerg. 1858, 1–11 (2017).
Sluchanko, N. N., Slonimskiy, Y. B., Moldenhauer, M., Friedrich, T. & Maksimov, E. G. Deletion of the short N-terminal extension in OCP reveals the main site for FRP binding. FEBS Lett. 591, 1667–1676 (2017).
Lu, Y. et al. Native mass spectrometry analysis of oligomerization states of fluorescence recovery protein and orange carotenoid protein: Two proteins involved in the cyanobacterial photoprotection cycle. Biochemistry 56, 160–166 (2017).
Moldenhauer, M. et al. Interaction of the signaling state analog and the apoprotein form of the orange carotenoid protein with the fluorescence recovery protein. Photosynth. Res. 135, 125–139 (2018).
Thurotte, A. et al. The cyanobacterial fluorescence recovery protein has two distinct activities: orange carotenoid protein amino acids involved in FRP interaction. Biochim. Biophys. Acta Bioenerg. 1858, 308–317 (2017).
Bao, H. et al. Additional families of orange carotenoid proteins in the photoprotective system of cyanobacteria. Nat. Plants 3, 17089 (2017).
Lechno-Yossef, S., Melnicki, M. R., Bao, H., Montgomery, B. L. & Kerfeld, C. A. Synthetic OCP heterodimers are photoactive and recapitulate the fusion of two primitive carotenoproteins in the evolution of cyanobacterial photoprotection. Plant J. 91, 646–656 (2017).
Melnicki, M. R. et al. Structure, diversity, and evolution of a new family of soluble carotenoid-binding proteins in cyanobacteria. Mol. Plant 9, 1379–1394 (2016).
Muzzopappa, F. et al. The paralogs to the C-terminal domain of the cyanobacterial OCP are carotenoid donors to HCPs. Plant Physiol. 175, 1283–1303 (2017).
Kerfeld, C. A. Structure and function of the water-soluble carotenoid-binding proteins of cyanobacteria. Photosynth. Res. 81, 215–225 (2004).
Wilson, A. et al. Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria. J. Biol. Chem. 285, 18364–18375 (2010).
Maksimov, E. G. et al. The unique protein-to-protein carotenoid transfer mechanism. Biophys. J. 113, 402–414 (2017).
Moldenhauer, M. et al. Assembly of photoactive orange carotenoid protein from its domains unravels a carotenoid shuttle mechanism. Photosynth. Res. 133, 327–341 (2017).
Jallet, D. et al. Specificity of the cyanobacterial orange carotenoid protein: influences of orange carotenoid protein and phycobilisome structures. Plant Physiol. 164, 790–804 (2014).
Bernát, G. et al. Unique properties vs. common themes: the atypical cyanobacterium Gloeobacter violaceus PCC 7421 is capable of state transitions and blue-light-induced fluorescence quenching. Plant Cell Physiol. 53, 528–542 (2012).
Harris, D. et al. Structural rearrangements in the C-terminal domain homolog of orange carotenoid protein are crucial for carotenoid transfer. Commun. Biol. 1, 125 (2018).
Slonimskiy, Y. B. et al. Light-controlled carotenoid transfer between water-soluble proteins related to cyanobacterial photoprotection. FEBS J. 286, 1908–1924 (2019).
Maksimov, E. G. et al. Fluorescent labeling preserving OCP photoactivity reveals its reorganization during the photocycle. Biophys. J. 112, 827 (2017).
Maksimov, E. G. et al. The photocycle of orange carotenoid protein conceals distinct intermediates and asynchronous changes in the carotenoid and protein components. Sci. Rep. 7, 15548 (2017).
López-Igual, R. et al. Different functions of the paralogs to the N-terminal domain of the orange carotenoid protein in the cyanobacterium Anabaena sp. PCC 7120. Plant Physiol. 171, 1852–1866 (2016).
Wen, Y. et al. Orange and red carotenoid proteins are involved in the adaptation of the terrestrial cyanobacterium Nostoc flagelliforme to desiccation. Photosynth. Res. 1, 1–11 (2019).
Harris, D. et al. Orange carotenoid protein burrows into the phycobilisome to provide photoprotection. Proc. Natl Acad. Sci. USA 113, E1655–E1662 (2016).
Sluchanko, N. N. et al. OCP–FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria. Nat. Commun. 9, 1–15 (2018).
Bourcier De Carbon, C., Thurotte, A., Wilson, A., Perreau, F. & Kirilovsky, D. Biosynthesis of soluble carotenoid holoproteins in Escherichia coli. Sci. Rep. 5, 9085 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539 (2011).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Cock, P. J. A. et al. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Wheeler, T. J., Clements, J. & Finn, R. D. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden markov models. BMC Bioinformatics 15, 7 (2014).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Berendsen, H. J. C., van der Spoel, D. & van Drunen, R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995).
Acknowledgements
The authors thank S. Cot for technical help, F. Andre for helping with the phylogenetic analysis, L. Tabares for assisting with molecular dynamics simulations and M. Guillaume Sarrailhe for helping in the construction of the three OCP mutants. This work was supported by grants from the Agence Nationale de la Recherche (RECYFUEL project (grant no. ANR-16-CE05- 0026)) and from the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 675006 (SE2B)). F.M.’s salary was financed by the European Union’s Horizon 2020 (project no. SE2B). The research was also supported by the Centre National de la Recherche Scientifique and the Commissariat à l’Energie Atomique. The French Infrastructure for Integrated Structural Biology (grant no. ANR-10-INBS-05) also partially supported this research.
Author information
Authors and Affiliations
Contributions
F.M. performed all the OCP characterization experiments and constructed some of the mutants. A.W. constructed almost all the OCP mutants. D.K. conceived the project, designed and supervised most of the experiments and analysed the data. The article was written by F.M. and D.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Roberto Bassi and Conrad Mullineaux and other, anonymous, reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Table 1.
Supplementary Dataset
Sequences of OCPs used for construction of the phylogenetic tree.
Rights and permissions
About this article
Cite this article
Muzzopappa, F., Wilson, A. & Kirilovsky, D. Interdomain interactions reveal the molecular evolution of the orange carotenoid protein. Nat. Plants 5, 1076–1086 (2019). https://doi.org/10.1038/s41477-019-0514-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0514-9
This article is cited by
-
Accidental surface compatibility paved the way for a new molecular interaction
Nature Ecology & Evolution (2023)
-
Fortuitously compatible protein surfaces primed allosteric control in cyanobacterial photoprotection
Nature Ecology & Evolution (2023)
-
Cyclophosphamide treatment modifies the thermal stability of profilin bound monomeric and leiomodin2 bound filamentous actin
Journal of Thermal Analysis and Calorimetry (2023)
-
Dose-dependent effect of cyclophosphamide treatment on actin
Journal of Thermal Analysis and Calorimetry (2022)
-
Alterations of inter-domain flexibility in actin monomers during cyclophosphamide treatment
Journal of Thermal Analysis and Calorimetry (2022)