Abstract
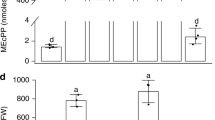
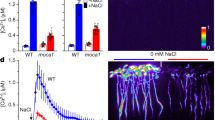
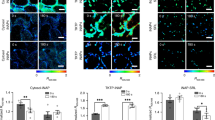
The signalling lipid phosphatidic acid (PA) is involved in regulating various fundamental biological processes in plants. However, the mechanisms of PA action remain poorly understood because currently available methods for monitoring PA fail to determine the precise spatio-temporal dynamics of this messenger in living cells and tissues of plants. Here, we have developed PAleon, a PA-specific optogenetic biosensor that reports the concentration and dynamics of bioactive PA at the plasma membrane based on Förster resonance energy transfer (FRET). PAleon was sensitive enough to monitor physiological concentrations of PA in living cells and to visualize PA dynamics at subcellular resolution in tissues when they were challenged with abscisic acid (ABA) and salt stress. PAleon bioimaging revealed kinetics and tissue specificity of salt stress-triggered PA accumulation. Compared with wild-type Arabidopsis, the pldα1 mutant lacking phospholipase Dα1 (PLDα1) for PA generation showed delayed and reduced PA accumulation. Comparative analysis of wild type and pldα1 mutant indicated that cellular pH-modulated PA interaction with target proteins and PLD/PA-mediated salt tolerance. Application of the PA biosensor PAleon uncovered specific spatio-temporal PA dynamics in plant tissues. Our findings suggest that PA signalling integrates with cellular pH dynamics to mediate plant response to salt stress.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the paper and its Supplementary Information or from the corresponding authors upon request.
References
Pokotylo, I., Kravets, V., Martinecc, J. & Ruelland, E. The phosphatidic acid paradox: too many actions for one molecule class? Lessons from plants. Prog. Lipid Res. 71, 43–53 (2018).
Young, B. P. et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329, 1085–1088 (2010).
Wang, X., Su, Y., Liu, Y., Kim, S. C. & Fanella, B. Signaling and Communication in Plants (ed. Wang, X.) 69–92 (Springer, 2014).
Kassas, N. et al. Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. J. Biol. Chem. 292, 4266–4279 (2017).
Jacob, T., Ritchie, S., Assmann, S. & Gilroy, S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl Acad. Sci. USA 96, 12192–12197 (1999).
Mishra, G., Zhang, W., Deng, F., Zhao, J. & Wang, X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312, 264–266 (2006).
Zhang, Y. et al. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377 (2009).
Zhang, Q. et al. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576 (2012).
Galvan-Ampudia, C. S. et al. Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23, 2044–2050 (2013).
Wang, P. et al. Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED 2 auxin efflux transporter in response to salt stress. Plant Cell 31, 250–271 (2019).
McLoughlin, F. et al. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72, 436–449 (2012).
Testerink, C. & Munnik, T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361 (2011).
Welti, R. et al. Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002 (2002).
Shadyro, O., Yurkova, I., Kisel, M., Brede, O. & Arnhold, J. Formation of phosphatidic acid, ceramide, and diglyceride on radiolysis of lipids: identification by MALDI-TOF mass spectrometry. Free Radic. Biol. Med. 36, 1612–1624 (2004).
Bargmann, B. O. R. et al. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 50, 78–89 (2009).
An, N., Rudge, S. A., Zhang, Q. & Wakelam, M. J. Using lipidomics analysis to determine signalling and metabolic changes in cells. Curr. Opin. Biotech. 43, 96–103 (2017).
Nakanishi, H., Santos, P. D. L. & Neiman, A. M. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15, 1802–1815 (2004).
Rizzo, M. A., Shome, K., Watkins, S. C. & Romero, G. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J. Biol. Chem. 275, 23911–23918 (2000).
Potocký, M. et al. Live-cell imaging of phosphatidic acid dynamics in pollen tubes visualized by Spo20p-derived biosensor. New Phytol. 203, 483–494 (2014).
Platre, M. P. et al. A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev. Cell 45, 465–480 (2018).
Tavare, J. M., Fletcher, L. M. & Welsh, G. I. Using green fluorescent protein to study intracellular signaling. J. Endocrinol. 170, 297–306 (2001).
Wang, X., Devaiah, S. P., Zhang, W. & Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278 (2006).
Zhang, W., Qin, C., Zhao, J. & Wang, X. Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl Acad. Sci. USA 101, 9508–9513 (2004).
Nishioka, T., Frohman, M. A., Matsuda, M. & Kiyokawa, E. Heterogeneity of phosphatidic acid levels and distribution at the plasma membrane in living cells as visualized by a Förster resonance energy transfer (FRET) biosensor. J. Biol. Chem. 285, 35979–35987 (2010).
Yu, L. et al. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773 (2010).
Devaiah, S. P. et al. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dα1 knockout mutant. Phytochemistry 67, 1907–1924 (2006).
Ni, M. et al. Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7, 661–676 (1995).
Simon, M. L. et al. A PtdIns(4)P-deriven electrostatic field controls cell membrane identity and signaling in plants. Nat. Plants 2, 16089–16098 (2016).
Waadt, R. et al. FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3, e01739 (2014).
Behera, S. et al. Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30, 2704–2719 (2018).
Wilkins, K. A. et al. Self-incompatibility-induced programmed cell death in field poppy pollen involves dramatic acidification of the incompatible pollen tube cytosol. Plant Physiol. 167, 766–779 (2015).
Darwish, E., Testerink, C., Khalil, M., Elshihy, O. & Munnik, T. Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell Physiol. 50, 986–997 (2009).
Jones, A. M. et al. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3, e01741 (2014).
Petersen, E. N., Chung, H. W., Nayebosadri, A. & Hansen, S. B. Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat. Commun. 7, 13873 (2016).
Basu, D. & Haswell, E. S. Plant mechanosensitive ion channels: an ocean of possibilities. Curr. Opin. Plant Biol. 40, 43–48 (2017).
Zhao, J. & Wang, X. Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J. Biol. Chem. 279, 1794–1800 (2004).
Martinière, A. et al. Uncovering pH at both sides of the root plasma membrane interface using noninvasive imaging. Proc. Natl Acad. Sci. USA 115, 6488–6493 (2018).
Rocks, O., Peyker, A. & Bastiaens, P. I. H. Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Curr. Opin. Cell Biol. 18, 351–357 (2006).
Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008).
Yang., Y. & Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539 (2018).
Gevaudant, F. et al. Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol. 144, 1763–1776 (2007).
Wang, X. Multiple forms of phospholipase D in plants: the gene family, catalytic and regulatory properties, and cellular functions. Prog. Lipid Res. 39, 109–149 (2000).
Hong, Y., Pan, X., Welti, R. & Wang, X. Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20, 803–816 (2008).
Krebs, M. et al. FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J. 69, 181–192 (2012).
Acknowledgements
We thank E. Kiyokawa (Department of Pathology and Biology of Diseases, Graduate School of Medicine, Kyoto University) for the generous gift of the Pii vector. We thank J. Chen and X. Liu for their assistance in using confocals. The research was supported by grants from National Natural Science Foundation of China (grant nos. 31770294 and 31570270) to W.Z. and from the Deutsche Forschungsgemeinschaft (grant no. DFG, KU931/14-1).
Author information
Authors and Affiliations
Contributions
W.L. and W.Z. designed experiments, analysed data and wrote the manuscript. W.L. performed most of the experiments and prepared the data. T.S. and L.W. helped with the experiments. J.K., L.W. and X.W. discussed the data and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and an additional figure.
Rights and permissions
About this article
Cite this article
Li, W., Song, T., Wallrad, L. et al. Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat. Plants 5, 1012–1021 (2019). https://doi.org/10.1038/s41477-019-0497-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0497-6
This article is cited by
-
Phosphatidic acid priming-enhanced heat tolerance in tall fescue (Festuca arundinacea) involves lipidomic reprogramming of lipids for membrane stability and stress signaling
Plant Growth Regulation (2023)
-
Identification, evolution, expression analysis of phospholipase D (PLD) gene family in tea (Camellia sinensis)
Physiology and Molecular Biology of Plants (2021)
-
Overexpression of phosphatidylserine synthase IbPSS1 affords cellular Na+ homeostasis and salt tolerance by activating plasma membrane Na+/H+ antiport activity in sweet potato roots
Horticulture Research (2020)
-
Lipids light up in plant membranes
Nature Plants (2019)