Abstract
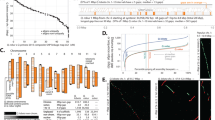
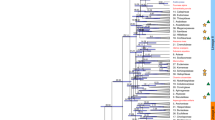
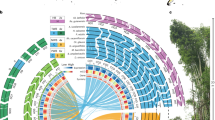
Comparative genomics can unravel the genetic basis of species differences; however, successful reports on quantitative traits are still scarce. Here we present genome assemblies of 31 so-far unassembled Brassicaceae plant species and combine them with 16 previously published assemblies to establish the Brassicaceae Diversity Panel. Using a new interspecies association strategy for quantitative traits, we found a so-far unknown association between the unexpectedly high variation in CG to TG substitution rates in genes and the absence of CHROMOMETHYLASE3 (CMT3) orthologues. Low substitution rates were associated with the loss of CMT3, while species with conserved CMT3 orthologues showed high substitution rates. Species without CMT3 also lacked gene-body methylation (gbM), suggesting an evolutionary trade-off between the unknown function of gbM and low substitution rates in Brassicaceae, possibly due to low mutability of non-methylated cytosines.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All sequencing data, assemblies and gene annotations generated in this project can be found in the European Nucleotide Archive under the project accession number PRJEB26555.
Code availability
The code used to generate associations, the species tree, and the genotypic and phenotypic data can be accessed at https://github.com/schneebergerlab/Scripts_supporting_Kiefer_Willing_et_al_2019.
References
Huynen, M., Dandekar, T. & Bork, P. Differential genome analysis applied to the species-specific features of Helicobacter pylori. FEBS Lett. 426, 1–5 (1998).
Gaasterland, T. & Ragan, M. A. Constructing multigenome views of whole microbial genomes. Microb. Comp. Genomics 3, 177–192 (1998).
Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA 96, 4285–4288 (1999).
Aravind, L., Watanabe, H., Lipman, D. J. & Koonin, E. V. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA 97, 11319–11324 (2000).
Hiller, M. et al. A ‘forward genomics’ approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2, 817–823 (2012).
Prudent, X., Parra, G., Schwede, P., Roscito, J. G. & Hiller, M. Controlling for phylogenetic relatedness and evolutionary rates improves the discovery of associations between species’ phenotypic and genomic differences. Mol. Biol. Evol. 33, 2135–2150 (2016).
Delaux, P.-M. et al. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 10, e1004487 (2014).
Griesmann, M. et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361, eaat1743 (2018).
Zhang, G. et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320 (2014).
Burga, A. et al. A genetic signature of the evolution of loss of flight in the Galapagos cormorant. Science 356, eaal3345 (2017).
Pease, J. B., Haak, D. C., Hahn, M. W. & Moyle, L. C. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14, e1002379 (2016).
Vlad, D. et al. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343, 780–783 (2014).
Bewick, A. J. et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl Acad. Sci. USA 113, 9111–9116 (2016).
Willing, E.-M. et al. UVR2 ensures transgenerational genome stability under simulated natural UV-B in Arabidopsis thaliana. Nat. Commun. 7, 13522 (2016).
Exposito-Alonso, M. et al. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 14, e1007155 (2018).
Ossowski, S. et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94 (2010).
Willing, E.-M. et al. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat. Plants 1, 14023 (2015).
Briskine, R. V. et al. Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Mol. Ecol. Resour. 17, 1025–1036 (2017).
The Brassica rapa Genome Sequencing Project Consortium et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039 (2011).
Yang, J. et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48, 1225–1232 (2016).
Chalhoub, B. et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014).
Jiao, W.-B. et al. Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 27, 778–786 (2017).
Haudry, A. et al. An atlas of over 90,000 conserved non-coding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45, 891–898 (2013).
Moghe, G. D. et al. Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. Plant Cell 26, 1925–1937 (2014).
Dassanayake, M. et al. The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913–918 (2011).
Yang, R. et al. The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 4, 46 (2013).
Slotte, T. et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45, 831–835 (2013).
Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Hu, T. T. et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481 (2011).
Gan, X. et al. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants 2, 16167 (2016).
Lee, C.-R. et al. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat. Ecol. Evol. 1, 0119 (2017).
Kiefer, M. et al. BrassiBase: introduction to a novel knowledge database on Brassicaceae evolution. Plant Cell Physiol. 55, e3 (2014).
Koch, M. A., German, D. A., Kiefer, M. & Franzke, A. Database taxonomics as key to modern plant biology. Trends Plant Sci. 23, 4–6 (2018).
Guo, X. et al. Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics 18, 176 (2017).
Hohmann, N., Wolf, E. M., Lysak, M. A. & Koch, M. A. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27, 2770–2784 (2015).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015).
Duarte, J. M. et al. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 10, 61 (2010).
Zhang, N., Zeng, L., Shan, H. & Ma, H. Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 195, 923–937 (2012).
Huang, C.-H. et al. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 33, 394–412 (2016).
Nikolov, L. A. et al. Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 222, 1638–1651 (2019).
Kagale, S. et al. Polyploid evolution of the Brassicaceae during the Cenozoic Era. Plant Cell 26, 2777–2791 (2014).
Mandáková, T., Li, Z., Barker, M. S. & Lysak, M. A. Diverse genome organization following 13 independent mesopolyploid events in Brassicaceae contrasts with convergent patterns of gene retention. Plant J. 91, 3–21 (2017).
Li, Z. et al. Early genome duplications in conifers and other seed plants. Sci. Adv. 1, e1501084 (2015).
Pagel, M. Inferring evolutionary processes from phylogenies. Zool. Scr. 26, 331–348 (1997).
Pagel, M. Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999).
Grafen, A. The phylogenetic regression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326, 119–157 (1989).
Felsenstein, J. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 25, 471–492 (1973).
Felsenstein, J. Phylogenies and quantitative characters. Annu. Rev. Ecol. Syst. 19, 445–471 (1988).
Vuolo, F. et al. Coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev. 30, 2370–2375 (2016).
Harris, K. & Pritchard, J. K. Rapid evolution of the human mutation spectrum. eLife 6, e24284 (2017).
Harris, K. Evidence for recent, population-specific evolution of the human mutation rate. Proc. Natl Acad. Sci. USA 112, 3439–3444 (2015).
Lynch, M. Evolution of the mutation rate. Trends Genet. 26, 345–352 (2010).
Mathieson, I. & Reich, D. Differences in the rare variant spectrum among human populations. PLoS Genet. 13, e1006581 (2017).
Gou, L., Bloom, J. S. & Kruglyak, L. The genetic basis of mutation rate variation in yeast. Genetics 211, 731–740 (2019).
Pang, Q., Hays, J. B., Rajagopal, I. & Schaefer, T. S. Selection of Arabidopsis cDNAs that partially correct phenotypes of Escherichia coli DNA-damage-sensitive mutants and analysis of two plant cDNAs that appear to express UV-specific dark repair activities. Plant Mol. Biol. 22, 411–426 (1993).
Du, J., Johnson, L. M., Jacobsen, S. E. & Patel, D. J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 (2015).
Becker, C. et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249 (2011).
Niederhuth, C. E. et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17, 194 (2016).
Bewick, A. J. et al. The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18, 65 (2017).
Zhao, T. & Schranz, M. E. Network-based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes. Proc. Natl Acad. Sci. USA 116, 2165–2174 (2019).
Albalat, R. & Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 17, 379–391 (2016).
Chin, C.-S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Pryszcz, L. P. & Gabaldón, T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44, e113 (2016).
Sun, H., Ding, J., Piednoël, M., Schneeberger, K. & Birol, I. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics 34, 550–557 (2018).
Keller, O., Odronitz, F., Stanke, M., Kollmar, M. & Waack, S. Scipio: using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinformatics 9, 278 (2008).
Stanke, M. & Morgenstern, B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 (2005).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Korf, I. Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Chen, J., Hu, Q., Zhang, Y., Lu, C. & Kuang, H. P-MITE: a database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 42, D1176–D1181 (2014).
Macas, J., Mészáros, T. & Nouzová, M. PlantSat: a specialized database for plant satellite repeats. Bioinformatics 18, 28–35 (2002).
Ouyang, S. & Buell, C. R. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 32, D360–D363 (2004).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Nussbaumer, T. et al. MIPS PlantsDB: a database framework for comparative plant genome research. Nucleic Acids Res. 41, D1144–D1151 (2013).
Goubert, C. et al. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Biol. Evol. 7, 1192–1205 (2015).
Guo, W. et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14, 774 (2013).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics https://doi.org/10.1093/bioinformatics/btz305 (2019).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Lanfear, R., Calcott, B., Ho, S. Y. W. & Guindon, S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701 (2012).
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T. & Calcott, B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773 (2017).
Zhang, C., Rabiee, M., Sayyari, E. & Mirarab, S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19, 153 (2018).
Bouckaert, R. et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15, e1006650 (2019).
Vanneste, K., Van de Peer, Y. & Maere, S. Inference of genome duplications from age distributions revisited. Mol. Biol. Evol. 30, 177–190 (2013).
Vanneste, K., Baele, G., Maere, S. & Van de Peer, Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res. 24, 1334–1347 (2014).
Seymour, D. K., Koenig, D., Hagmann, J., Becker, C. & Weigel, D. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 10, e1004785 (2014).
Orme, D. The caper package: comparative analysis of phylogenetics and evolution in R (CRAN, 2018); https://cran.r-project.org/web/packages/caper/vignettes/caper.pdf
Acknowledgements
We would like to thank Bruno Hüttel (Max Planck Genome Centre, Cologne, Germany). This work was supported by the ERC STG Grant INTERACT 802629 (K.S.). Work on BrassiBase was funded within the framework of DFG-SPP 1529 (KO2302-13-1/2) (M.A.K.).
Author information
Authors and Affiliations
Contributions
K.S., M.A.K., E.-M.W. and C.K. designed research. C.K., E.-M.W., W.-B.J., H.S., M.P. and K.S. analysed the data. C.K., U.H. and B.H. performed the plant work. K.S. wrote the paper with the help of all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Materials and Methods and Supplementary Figs. 1–16.
Supplementary Table 1
Selected species, generated sequencing data and assembly quality.
Supplementary Table 2
Meso-polyploidization prediction (lower part) trained on genomes with known meso-polyploidizations.
Supplementary Table 3
MAPS analysis of species with putatively shared WGD events.
Supplementary Table 4
Leaf shape complexity values.
Supplementary Table 5
Orthologous groups (OG) with highly significant associations to leaf shape complexity.
Rights and permissions
About this article
Cite this article
Kiefer, C., Willing, EM., Jiao, WB. et al. Interspecies association mapping links reduced CG to TG substitution rates to the loss of gene-body methylation. Nat. Plants 5, 846–855 (2019). https://doi.org/10.1038/s41477-019-0486-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0486-9
This article is cited by
-
Analysis of flavonol regulator evolution in the Brassicaceae reveals MYB12, MYB111 and MYB21 duplications and MYB11 and MYB24 gene loss
BMC Genomics (2022)
-
Molecular properties of epimutation hotspots
Nature Plants (2022)
-
Heterochromatin is a quantitative trait associated with spontaneous epiallele formation
Nature Communications (2021)
-
Nested whole-genome duplications coincide with diversification and high morphological disparity in Brassicaceae
Nature Communications (2020)
-
DNA methylation makes mutational history
Nature Plants (2019)