Abstract
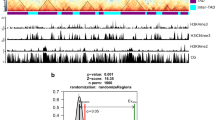
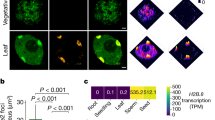
Chromatin conformation capture (3C)1 and high-throughput 3C (Hi-C)2 assays allow the study of three-dimensional (3D) genome structures in cell populations or tissues, based on average proximities of folded DNA. However, differences between cells can be observed only by single-cell measurements that avoid ensemble averaging3,4,5. To study 3D chromatin organization and dynamics before and after fertilization in flowering plants, we analysed the 3D genomes of rice eggs, sperm cells, unicellular zygotes and shoot mesophyll cells. We show that chromatin architectures of rice eggs and sperm cells are comparable to those of mesophyll cells and are reorganized after fertilization. The rice single-cell 3D genomes display specific features of chromosome compartments and telomere/centromere configuration compared to those in mammalian single cells. Active and silent chromatin domains combine to form multiple foci in the nuclear space. Notably, the 3D genomes of the eggs and unicellular zygotes contain a compact silent centre (CSC) that is absent in sperm cells. CSC appears to be reorganized after fertilization, and may be involved in the regulation of zygotic genome activation (ZGA). Our results reveal specific 3D genome features of plant gametes and the unicellular zygote, and provide a spatial chromatin basis for ZGA and epigenetic regulation in plants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data in support of the findings of this study, including the raw data, the output ncc files of Nuc_processing software and the final programme database files for 3D viewing of the genome in PyMOL, are deposited in the Gene Expression Omnibus (No. GSE123109) and are available from the corresponding author upon request.
References
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Tan, L., Xing, D., Chang, C. H., Li, H. & Xie, X. S. Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018).
Nagano, T. et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017).
Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Liu, C., Cheng, Y. J., Wang, J. W. & Weigel, D. Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat. Plants 3, 742–748 (2017).
Dong, P. et al. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol. Plant 10, 1497–1509 (2017).
Wang, M. et al. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat. Plants 4, 90–97 (2018).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Dong, F. & Jiang, J. Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res. 6, 551–558 (1998).
Cowan, C. R., Carlton, P. M. & Cande, W. Z. The polar arrangement of telomeres in interphase and meiosis. Rabl organization and the bouquet. Plant Physiol. 125, 532–538 (2001).
Flyamer, I. M. et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017).
Zhao, P. & Sun, M. X. The maternal-to-zygotic transition in higher plants: available approaches, critical limitations, and technical requirements. Curr. Top. Dev. Biol. 113, 373–398 (2015).
Chen, J. et al. Zygotic genome activation occurs shortly after fertilization in maize. Plant Cell 29, 2106–2125 (2017).
Liu, H. & Nonomura, K. I. A wide reprogramming of histone H3 modifications during male meiosis I in rice is dependent on the Argonaute protein MEL1. J. Cell. Sci. 129, 3553–3561 (2016).
Feng, S. et al. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 55, 694–707 (2014).
Grob, S., Schmid, M. W. & Grossniklaus, U. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell 55, 678–693 (2014).
Anderson, S. N. et al. The zygotic transition Is initiated in unicellular plant zygotes with asymmetric activation of parental genomes. Dev. Cell 43, 349–358 (2017).
Jukam, D., Shariati, S. A. M. & Skotheim, J. M. Zygotic genome activation in vertebrates. Dev. Cell 42, 316–332 (2017).
Lee, M. T., Bonneau, A. R. & Giraldez, A. J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell. Dev. Biol. 30, 581–613 (2014).
Dong, Q. et al. Genome-wide Hi-C analysis reveals extensive hierarchical chromatin interactions in rice. Plant J. 94, 1141–1156 (2018).
Acknowledgements
We thank X. Li and C. Luo for help with single-cell isolation. This work was supported by grants from the National Key Research and Development Programme of China (No. 2016YFD0100802), the National Natural Science Foundation of China (No. 31730049), Fundamental Research Funds for the Central Universities (No. 2662015PY228) and the National Postdoctoral Programme for Innovative Talents.
Author information
Authors and Affiliations
Contributions
S.Z. performed most of the experiments, analysed the data and participated in writing of the paper. W.J. and Y.Z. participated in the experimental set-up of single-cell isolation. D.-X.Z. supervised the project, analysed the data and wrote the paper, with input from S.Z.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Plants thanks Thomas Dresselhaus, Chang Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Supplementary Methods and Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Zhou, S., Jiang, W., Zhao, Y. et al. Single-cell three-dimensional genome structures of rice gametes and unicellular zygotes. Nat. Plants 5, 795–800 (2019). https://doi.org/10.1038/s41477-019-0471-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0471-3
This article is cited by
-
DNA methylation remodeling and the functional implication during male gametogenesis in rice
Genome Biology (2024)
-
3D organization of regulatory elements for transcriptional regulation in Arabidopsis
Genome Biology (2023)
-
Application of single-cell multi-omics approaches in horticulture research
Molecular Horticulture (2023)
-
Paternal DNA methylation is remodeled to maternal levels in rice zygote
Nature Communications (2023)
-
HSFA1a modulates plant heat stress responses and alters the 3D chromatin organization of enhancer-promoter interactions
Nature Communications (2023)