Abstract
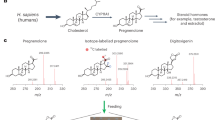
Brassinosteroids (BRs) are essential plant steroid hormones that regulate plant growth and development1. The most potent BR, brassinolide, is produced by addition of many oxygen atoms to campesterol by several cytochrome P450 monooxygenases (CYPs). CYP90B1 (also known as DWF4) catalyses the 22(S)-hydroxylation of campesterol and is the first and rate-limiting enzyme at the branch point of the biosynthetic pathway from sterols to BRs2. Here we show the crystal structure of Arabidopsis thaliana CYP90B1 complexed with cholesterol as a substrate. The substrate-binding conformation explains the stereoselective introduction of a hydroxy group at the 22S position, facilitating hydrogen bonding of brassinolide with the BR receptor3,4,5. We also determined the crystal structures of CYP90B1 complexed with uniconazole6,7 or brassinazole8, which inhibit BR biosynthesis. The two inhibitors are structurally similar; however, their binding conformations are unexpectedly different. The shape and volume of the active site pocket varies depending on which inhibitor or substrate is bound. These crystal structures of plant CYPs that function as membrane-anchored enzymes and exhibit structural plasticity can inform design of novel inhibitors targeting plant membrane-bound CYPs, including those involved in BR biosynthesis, which could then be used as plant growth regulators and agrochemicals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank under accession codes 6A15 (cholesterol-bound form), 6A16 (uniconazole-bound form), 6A17 (brassinazole-bound form) and 6A18 (1,6-hexanediol-bound form). All other data are available from the corresponding author upon reasonable request.
References
Grove, M. D. et al. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281, 216–217 (1979).
Fujita, S. et al. Arabidopsis CYP90B1 catalyses the early C‐22 hydroxylation of C27, C28 and C29 sterols. Plant J. 45, 765–774 (2006).
Santiago, J., Henzler, C. & Hothorn, M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889–892 (2013).
She, J. et al. Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476 (2011).
Hothorn, M. et al. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471 (2011).
Saito, S. et al. A plant growth retardant, uniconazole, is a potent inhibitor of ABA catabolism in Arabidopsis. Biosci. Biotechnol. Biochem. 70, 1731–1739 (2006).
Iwasaki, T. & Shibaoka, H. Brassinosteroids act as regulators of tracheary-element differentiation in isolated Zinnia mesophyll cells. Plant Cell Physiol. 32, 1007–1014 (1991).
Asami, T. et al. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123, 93–99 (2000).
Kühnel, K. et al. Crystal structures of substrate-free and retinoic acid-bound cyanobacterial cytochrome P450 CYP120A1. Biochemistry 47, 6552–6559 (2008).
Strushkevich, N. et al. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc. Natl Acad. Sci. USA 108, 15535–15535 (2011).
Mast, N. et al. Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain. Proc. Natl Acad. Sci. USA 105, 9546–9551 (2008).
Ouellet, H. et al. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 77, 730–742 (2010).
Strushkevich, N. et al. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc. Natl Acad. Sci. USA 108, 10139–10143 (2011).
Matsuoka, S. et al. Water‐mediated recognition of simple alkyl chains by heart‐type fatty‐acid‐binding protein†. Angew. Chem. Int. Ed. 54, 1508–1511 (2015).
Yamamoto, R., Demura, T. & Fukuda, H. Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant Cell Physiol. 38, 980–983 (1997).
Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 267, 83–90 (1992).
Asami, T. et al. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J. Biol. Chem. 276, 25687–25691 (2001).
Sekimata, K. et al. A specific brassinosteroid biosynthesis inhibitor, Brz001: evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta 213, 716–721 (2001).
Sekimata, K. et al. Brz220 interacts with DWF4, a cytochrome P450 monooxygenase in brassinosteroid biosynthesis, and exerts biological activity. Biosci. Biotechnol. Biochem. 72, 7–12 (2008).
Ohnishi, T. et al. CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 287, 31551–31560 (2012).
Ohnishi, T. et al. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18, 3275–3288 (2006).
Kwon, M. et al. A double mutant for the CYP85A1 and CYP85A2 genes of Arabidopsis exhibits a brassinosteroid dwarf phenotype. J. Plant. Biol. 48, 237–244 (2005).
Nielsen, K. A. & Møller, B. L. in Cytochrome P450 Structure, Mechanism, and Biochemistry (ed. Ortiz de Montellano, P. R.) 553–583 (Kluwer Academic/Plenum Publishers, 2005).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D 65, 582–601 (2009).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
The PyMOL Molecular Graphics System version 1.8 (Schrodinger, L. L. C., 2015).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Kusano, K., Waterman, M. R., Sakaguchi, M., Omura, T. & Kagawa, N. Protein synthesis inhibitors and ethanol selectively enhance heterologous expression of P450s and related proteins in Escherichia coli. Arch. Biochem. Biophys. 367, 129–136 (1999).
Saito, S. et al. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449 (2004).
Mizutani, M. & Ohta, D. Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116, 357–367 (1998).
Acknowledgements
We thank K. Okamoto for constructing various expression plasmids for CYP90B1 and for purification of this enzyme. We also thank the beamline staff of BL26B1, BL41XU and BL32XU at SPring-8 (Hyogo, Japan) for assistance with the experiments. This study was supported by JSPS KAKENHI grant number JP17H05444, JP17K05933 and JP18H03945. This study is partially supported by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Ministry of Education, Culture, Sports, Science (MEXT) and Japan Agency for Medical Research and Development (AMED) under grant number JP18am0101070.
Author information
Authors and Affiliations
Contributions
K.F., T.H. and M.K. performed enzyme purification and crystallography. K.F., T.H., M.M. and S.N. wrote the manuscript. B.W., H.J.L. and M.M. determined enzyme activities. All authors discussed the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Plants thanks Michael Hothorn and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–17 and Supplementary Tables 1–2.
Supplementary Video 1
Video showing cholesterol-binding mode.
Supplementary Video 2
Video showing uniconazole-binding mode.
Supplementary Video 3
Video showing brassinazole-binding mode
Rights and permissions
About this article
Cite this article
Fujiyama, K., Hino, T., Kanadani, M. et al. Structural insights into a key step of brassinosteroid biosynthesis and its inhibition. Nat. Plants 5, 589–594 (2019). https://doi.org/10.1038/s41477-019-0436-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0436-6
This article is cited by
-
Yeast-based system for in vivo evaluation of alleles of the anthocyanin production pathway
World Journal of Microbiology and Biotechnology (2023)
-
The protein conformational basis of isoflavone biosynthesis
Communications Biology (2022)
-
Metabolomic and transcriptomic analyses reveal the regulation of pigmentation in the purple variety of Dendrobium officinale
Scientific Reports (2020)