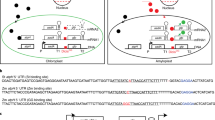
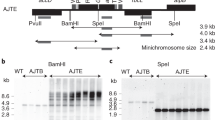
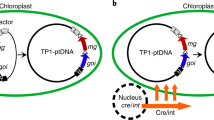
Abstract
The engineering of plant genomes presents exciting opportunities to modify agronomic traits and to produce high-value products in plants. Expression of foreign proteins from transgenes in the chloroplast genome offers advantages that include the capacity for prodigious protein output, the lack of transgene silencing and the ability to express multicomponent pathways from polycistronic mRNA. However, there remains a need for robust methods to regulate plastid transgene expression. We designed orthogonal activators that boost the expression of chloroplast transgenes harbouring cognate cis-elements. Our system exploits the programmable RNA sequence specificity of pentatricopeptide repeat proteins and their native functions as activators of chloroplast gene expression. When expressed from nuclear transgenes, the engineered proteins stimulate the expression of plastid transgenes by up to ~40-fold, with maximal protein abundance approaching that of Rubisco. This strategy provides a means to regulate and optimize the expression of foreign genes in chloroplasts and to avoid deleterious effects of their products on plant growth.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All essential data supporting the findings of this study are available within the paper and its Supplementary Information. Data for additional replicates are available from the corresponding author on request. Nucleotide sequences of the plastid transformation vectors are available under GenBank accessions MK482730 (pAI5), MK482731 (PQY1) and MK482728 (pQY3). Correspondence, requests for plasmids encoding the PPR10 variants and requests for transgenic plants expressing the PPR10 variants should be addressed to A.B. Requests for chloroplast transformation vectors and transplastomic plants lacking the PPR10 transgene should be addressed to P.M. Biological materials will be made available pending on the execution of a Materials Transfer Agreement with the University of Oregon and/or Rutgers University, as applicable.
References
Boynton, J. E. et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240, 1534–1538 (1988).
Svab, Z., Hajdukiewicz, P. & Maliga, P. Stable transformation of plastids in higher plants. Proc. Natl Acad. Sci. USA 87, 8526–8530 (1990).
Bock, R. Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66, 211–241 (2015).
Ahmad, N., Michoux, F., Lossl, A. G. & Nixon, P. J. Challenges and perspectives in commercializing plastid transformation technology. J. Exp. Bot. 67, 5945–5960 (2016).
Waheed, M. T., Ismail, H., Gottschamel, J., Mirza, B. & Lossl, A. G. Plastids: the green frontiers for vaccine production. Front. Plant Sci. 6, 1005 (2015).
Oey, M., Lohse, M., Kreikemeyer, B. & Bock, R. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 57, 436–445 (2009).
Fuentes, P. et al. A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. eLife 5, e13664 (2016).
Gnanasekaran, T. et al. Transfer of the cytochrome P450-dependent dhurrin pathway from Sorghum bicolor into Nicotiana tabacum chloroplasts for light-driven synthesis. J. Exp. Bot. 67, 2495–2506 (2016).
Harada, H. et al. Construction of transplastomic lettuce (Lactuca sativa) dominantly producing astaxanthin fatty acid esters and detailed chemical analysis of generated carotenoids. Transgenic Res. 23, 303–315 (2014).
Bohmert-Tatarev, K., McAvoy, S., Daughtry, S., Peoples, O. P. & Snell, K. D. High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol. 155, 1690–1708 (2011).
Diretto, G. et al. Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS ONE 2, e350 (2007).
Hanson, M. R., Lin, M. T., Carmo-Silva, A. E. & Parry, M. A. Towards engineering carboxysomes into C3 plants. Plant J. 87, 38–50 (2016).
Malhotra, K. et al. Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells. Mol. Plant 9, 1464–1477 (2016).
Lossl, A. et al. Inducible trans-activation of plastid transgenes: expression of the R. eutropha phb operon in transplastomic tobacco. Plant Cell Physiol. 46, 1462–1471 (2005).
Lu, Y., Rijzaani, H., Karcher, D., Ruf, S. & Bock, R. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc. Natl Acad. Sci. USA 110, E623–E632 (2013).
Scotti, N. & Cardi, T. Transgene-induced pleiotropic effects in transplastomic plants. Biotechnol. Lett. 36, 229–239 (2014).
Muhlbauer, S. K. & Koop, H. U. External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J. 43, 941–946 (2005).
Buhot, L., Horvath, E., Medgyesy, P. & Lerbs-Mache, S. Hybrid transcription system for controlled plastid transgene expression. Plant J. 46, 700–707 (2006).
Emadpour, M., Karcher, D. & Bock, R. Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res. 43, e66 (2015).
Barkan, A. & Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 (2014).
Small, I. & Peeters, N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 (2000).
Barkan, A. et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 (2012).
Shen, C. et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7, 11285 (2016).
Filipovska, A. & Rackham, O. Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol. Biosyst. 8, 699–708 (2012).
Kindgren, P., Yap, A., Bond, C. S. & Small, I. Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell 27, 403–416 (2015).
Colas des Francs-Small, C., Vincis Pereira Sanglard, L. & Small, I. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun. Biol. 1, 166 (2018).
Miranda, R. G., McDermott, J. J. & Barkan, A. RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR–RNA interactions. Nucleic Acids Res. 46, 2613–2623 (2018).
Pfalz, J., Bayraktar, O., Prikryl, J. & Barkan, A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28, 2042–2052 (2009).
Prikryl, J., Rojas, M., Schuster, G. & Barkan, A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA 108, 415–420 (2011).
Zoschke, R., Watkins, K. & Barkan, A. A rapid microarray-based ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25, 2265–2275 (2013).
Chotewutmontri, P. & Barkan, A. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 12, e1006106 (2016).
Miranda, R. G., Rojas, M., Montgomery, M. P., Gribbin, K. P. & Barkan, A. RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10. RNA 23, 586–599 (2017).
Schoffl, F., Raschke, E. & Nagao, R. T. The DNA sequence analysis of soybean heat-shock genes and identification of possible regulatory promoter elements. EMBO J. 3, 2491–2497 (1984).
Roslan, H. A. et al. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28, 225–235 (2001).
Erb, T. J. & Zarzycki, J. A short history of RubisCO: the rise and fall (?) of nature’s predominant CO2 fixing enzyme. Curr. Opin. Biotechnol. 49, 100–107 (2018).
Hui, M. P., Foley, P. L. & Belasco, J. G. Messenger RNA degradation in bacterial cells. Annu. Rev. Genet. 48, 537–559 (2014).
Germain, A., Hotto, A. M., Barkan, A. & Stern, D. B. RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 4, 295–316 (2013).
Luro, S., Germain, A., Sharwood, R. E. & Stern, D. B. RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res. 41, 9141–9151 (2013).
Beick, S., Schmitz-Linneweber, C., Williams-Carrier, R., Jensen, B. & Barkan, A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28, 5337–5347 (2008).
Hanson, M. R., Gray, B. N. & Ahner, B. A. Chloroplast transformation for engineering of photosynthesis. J. Exp. Bot. 64, 731–742 (2013).
Fuentes, P., Armarego-Marriott, T. & Bock, R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 49, 10–15 (2018).
Daniell, H., Chan, H. T. & Pasoreck, E. K. Vaccination via chloroplast genetics: affordable protein drugs for the prevention and treatment of inherited or infectious human diseases. Annu. Rev. Genet. 50, 595–618 (2016).
Zhou, F., Karcher, D. & Bock, R. Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 52, 961–972 (2007).
Hammani, K., Cook, W. & Barkan, A. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl Acad. Sci. USA 109, 5651–5656 (2012).
Legen, J. et al. Stabilization and translation of synthetic operon-derived mRNAs in chloroplasts by sequences representing PPR protein-binding sites. Plant J. 94, 8–21 (2018).
Ramundo, S. & Rochaix, J. D. Controlling expression of genes in the unicellular alga Chlamydomonas reinhardtii with a vitamin-repressible riboswitch. Methods Enzymol. 550, 267–281 (2015).
Boudreau, E., Nickelsen, J., Lemaire, S. D., Ossenbuhl, F. & Rochaix, J. D. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19, 3366–3376 (2000).
Kuchka, M. R., Goldschmidt-Clermont, M., van Dillewijn, J. & Rochaix, J. D. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58, 869–876 (1989).
Yu, Q., Barkan, A. & Maliga, P. Engineered RNA-binding protein for transgene activation in non-green plastids. Nat. Plants https://doi.org/10.1038/s41477-019-0413-0 (2019).
Kuroda, H. & Maliga, P. Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 125, 430–436 (2001).
Shinozaki, K. & Sugiura, M. Sequence of the intercistronic region between the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit and coupling factor beta subunit gene. Nucleic Acids Res. 10, 4923–4934 (1982).
Lutz, K. A., Svab, Z. & Maliga, P. Construction of marker-free transplastomic tobacco using the Cre-loxP site-specific recombination system. Nat. Protoc. 1, 900–910 (2006).
Earley, K. W. et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 (2006).
Werner, S., Breus, O., Symonenko, Y., Marillonnet, S. & Gleba, Y. High-level recombinant protein expression in transgenic plants by using a double-inducible viral vector. Proc. Natl Acad. Sci. USA 108, 14061–14066 (2011).
Acknowledgements
We are grateful to S. Belcher (University of Oregon) for assistance propagating tobacco and for help preparing figures, and to A. Ioannou and T. Tungsuchat Huang (Rutgers University) for their contributions to preparing plastid reporter constructs. We also appreciate the generous gifts of vectors from S. Strauss (Oregon State University) and D. Weigel (Max Planck Institute for Developmental Biology). This research was supported by the USDA NIFA Foundational Program Award number 2014-67013-21600 to A.B. and P.M.
Author information
Authors and Affiliations
Contributions
A.B. and P.M. conceived the study, designed the strategy and supervised the experiments. M.R., Q.Y. and R.W.-C. performed the experiments and interpreted the data. A.B. wrote the manuscript, which was edited by all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1 and 2.
Rights and permissions
About this article
Cite this article
Rojas, M., Yu, Q., Williams-Carrier, R. et al. Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat. Plants 5, 505–511 (2019). https://doi.org/10.1038/s41477-019-0412-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0412-1
This article is cited by
-
Expression strategies for the efficient synthesis of antimicrobial peptides in plastids
Nature Communications (2022)
-
U-to-C RNA editing by synthetic PPR-DYW proteins in bacteria and human culture cells
Communications Biology (2022)
-
Advances in plastid transformation for metabolic engineering in higher plants
aBIOTECH (2022)
-
A mitochondria-localized pentatricopeptide repeat protein is required to restore hau cytoplasmic male sterility in Brassica napus
Theoretical and Applied Genetics (2021)
-
Chloroplast (Cp) Transcriptome of P. davidiana Dode×P. bolleana Lauch provides insight into the Cp drought response and Populus Cp phylogeny
BMC Evolutionary Biology (2020)