Abstract
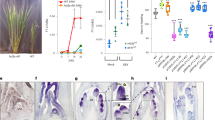
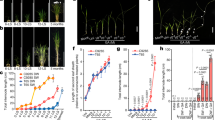
In several plant species, inflorescence formation is accompanied by stem elongation. Both processes are accelerated in rice upon perception of shortening days. Here, we show that PREMATURE INTERNODE ELONGATION 1 (PINE1), encoding a rice zinc-finger transcription factor, reduces the sensitivity of the stem to gibberellin (GA). The florigens reduce PINE1 expression to increase stem responsiveness to GA and promote flowering. These data indicate the existence of a regulatory network coordinating flowering and GA-dependent growth.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The RNA-seq data that support the findings of this study have been deposited in the GEO with the series record number GSE90493.
References
Brambilla, V. & Fornara, F. J. Integr. Plant Biol. 55, 410–418 (2013).
Zhao, J. et al. New Phytol. 208, 936–948 (2015).
Taoka, K. et al. Nature 476, 332–335 (2011).
Kobayashi, K. et al. Plant Cell 24, 1848–1859 (2012).
Tamaki, S. et al. Proc. Natl Acad. Sci. USA 112, E901–E910 (2015).
Hedden, P. & Sponsel, V. J. Plant Growth Regul. 34, 740–760 (2015).
Spielmeyer, W., Ellis, M. H. & Chandler, P. M. Proc. Natl Acad. Sci. USA 99, 9043–9048 (2002).
Harberd, N. P. et al. Nature 400, 256–261 (1999).
Sakamoto, T. et al. Nat. Biotechnol. 21, 909–913 (2003).
Hedden, P. & Thomas, S. G. Biochem. J. 444, 11–25 (2012).
Kaneko, M. et al. Plant J. 35, 104–115 (2003).
Furutani, I., Sukegawa, S. & Kyozuka, J. Plant J. 46, 503–511 (2006).
Doi, K. et al. Genes Dev. 18, 926–936 (2004).
Brambilla, V. et al. Plant Cell 29, 2801–2816 (2017).
Miao, J. et al. Cell Res. 23, 1233–1236 (2013).
Hoshikawa, K. The Growing Rice Plant (Nobunkyo, 1989).
Itoh, J. I., Kitano, H., Matsuoka, M. & Nagato, Y. Plant Cell 12, 2161–2174 (2000).
Sentoku, N. et al. Plant Cell 11, 1651–1664 (1999).
Tsuda, K., Ito, Y., Sato, Y. & Kurata, N. Plant Cell 23, 4368–4381 (2011).
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M. & Matsuoka, M. Plant Cell 14, 57–70 (2002).
Sakamoto, T. et al. Plant Physiol. 125, 1508–1516 (2001).
Nagai, K. et al. AoB Plants 6, plu028 (2014).
Gómez-Ariza, J. et al. J. Exp. Bot. 66, 2027–2039 (2015).
Fornara, F. et al. Plant Physiol. 135, 2207–2219 (2004).
Kim, D. et al. Genome Biol. 14, R36 (2013).
Langmead, B. & Salzberg, S. L. Nat. Methods 9, 357–359 (2012).
Anders, S., Pyl, P. T. & Huber, W. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. Bioinformatics 26, 139–140 (2010).
Gentleman, R. C. et al. Genome Biol. 5, R80 (2004).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Nat. Protoc. 8, 1551–1566 (2013).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Plant J. 6, 271–282 (1994).
Wild, M. et al. Dev. Cell 37, 190–200 (2016).
Acknowledgements
We are grateful to S. Tamaki for technical assistance with some experiments. The pACTIN vector was kindly provided by L. Dreni (IBMCP, CSIC, University of Valencia, Valencia) and the CRISPR–Cas9 vector by L.-J. Qu (Peking University and National Plant Gene Research Center, Beijing). We thank A. Costa and the NOLIMITS microscopy facility for technical support. This work was supported by an ERC Starting Grant (no. 260963) to F.F.
Author information
Authors and Affiliations
Contributions
J.G.-A., V.B., G.V., M.L., M.C., R.S. and F.G. performed the experiments, generated and analysed the transgenic plants and performed the hormone treatments. R.C. analysed the RNA-seq data. E.Carrera and I.L.D. quantified the bioactive GAs. E.Caporali performed the scanning electron microscopy. F.F. and J.G.-A. conceived the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information Nature Plants thanks Junko Kyozuka and other anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–10 and Supplementary Table 1.
Dataset S1
RNA-seq of Nipponbare and T65 shoot apices during meristem commitment.
Rights and permissions
About this article
Cite this article
Gómez-Ariza, J., Brambilla, V., Vicentini, G. et al. A transcription factor coordinating internode elongation and photoperiodic signals in rice. Nat. Plants 5, 358–362 (2019). https://doi.org/10.1038/s41477-019-0401-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0401-4
This article is cited by
-
Crop traits and production under drought
Nature Reviews Earth & Environment (2024)
-
Genome- and Transcriptome-wide Association Studies to Discover Candidate Genes for Diverse Root Phenotypes in Cultivated Rice
Rice (2023)
-
Genetic basis of geographical differentiation and breeding selection for wheat plant architecture traits
Genome Biology (2023)
-
Two florigens and a florigen-like protein form a triple regulatory module at the shoot apical meristem to promote reproductive transitions in rice
Nature Plants (2023)
-
Photoperiod and gravistimulation-associated Tiller Angle Control 1 modulates dynamic changes in rice plant architecture
Theoretical and Applied Genetics (2023)