Abstract
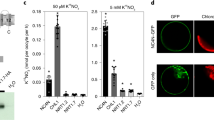
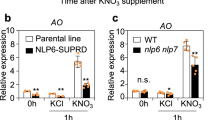
To ensure high crop yields in a sustainable manner, a comprehensive understanding of the control of nutrient acquisition is required. In particular, the signalling networks controlling the coordinated utilization of the two most highly demanded mineral nutrients, nitrogen and phosphorus, are of utmost importance. Here, we reveal a mechanism by which nitrate activates both phosphate and nitrate utilization in rice (Oryza sativa L.). We show that the nitrate sensor NRT1.1B interacts with a phosphate signalling repressor SPX4. Nitrate perception strengthens the NRT1.1B–SPX4 interaction and promotes the ubiquitination and degradation of SPX4 by recruiting NRT1.1B interacting protein 1 (NBIP1), an E3 ubiquitin ligase. This in turn allows the key transcription factor of phosphate signalling, PHR2, to translocate to the nucleus and initiate the transcription of phosphorus utilization genes. Interestingly, the central transcription factor of nitrate signalling, NLP3, is also under the control of SPX4. Thus, nitrate-triggered degradation of SPX4 activates both phosphate- and nitrate-responsive genes, implementing the coordinated utilization of nitrogen and phosphorus.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
Change history
24 April 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
15 May 2019
Owing to a technical fault, in the corrected version of the left panel of Fig. 4g, an extraneous vertical line appeared; this has now been removed.
References
Young, V. R. Nutritional balance studies: indicators of human requirements or of adaptive mechanisms? J. Nutr. 116, 700–703 (1986).
Hill, J. O. et al. Nutrient balance in humans: effects of diet composition. Am. J. Clin. Nutr. 54, 10–17 (1991).
Carreiro, A. L. et al. The macronutrients, appetite, and energy intake. Annu. Rev. Nutr. 36, 73–103 (2016).
Güsewell, S. N:P ratios in terrestrial plants: variation and functional significance. New Phytol. 164, 243–266 (2004).
Khan, F. et al. Effect of different levels of nitrogen and phosphorus on the phenology and yield of maize varieties. Am. J. Plant Sci. 5, 2582–2590 (2014).
Luo, X. et al. Nitrogen: phosphorous supply ratio and allometry in five alpine plant species. Ecol. Evol. 6, 8881–8892 (2016).
Güsewell, S. Responses of wetland graminoids to the relative supply of nitrogen and phosphorus. Plant Ecol. 176, 35–55 (2005).
Liu, K. H. & Tsay, Y. F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013 (2003).
Ho, C. H., Lin, S. H., Hu, H. C. & Tsay, Y. F. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 (2009).
Hu, B. et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838 (2015).
Marchive, C. et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4, 1713 (2013).
Liu, K. H. et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 545, 311–316 (2017).
Rubio, V. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133 (2001).
Lv, Q. et al. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26, 1586–1597 (2014).
Wild, R. et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990 (2016).
Puga, M. I. et al. SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in arabidopsis. Proc. Natl Acad. Sci. USA 111, 14947–14952 (2014).
Wang, Z. et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl Acad. Sci. USA 111, 14953–14958 (2014).
Kant, S., Peng, M. & Rothstein, S. J. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7, e1002021 (2011).
Medici, A. et al. Atnigt1/hrs1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6, 6274 (2015).
Kiba, T. et al. Repression of nitrogen-starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. Plant Cell 30, 925–945 (2018).
Maeda, Y. et al. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 9, 1376 (2018).
Sun, J. et al. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77 (2014).
Parker, J. L. & Newstead, S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72 (2014).
Chardin, C., Girin, T., Roudier, F., Meyer, C. & Krapp, A. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 65, 5577–5587 (2014).
Lopez-Arredondo, D. L., Leyva-Gonzalez, M. A., Alatorre-Cobos, F. & Herrera-Estrella, L. Biotechnology of nutrient uptake and assimilation in plants. Int. J. Dev. Biol. 57, 595–610 (2013).
Wang, R. et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136, 2512–2522 (2004).
Lin, S. I. et al. Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 51, 2119–2131 (2010).
Yue, W. et al. OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. 90, 1040–1051 (2017).
Lopez-Bucio, J., Cruz-Ramirez, A. & Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287 (2003).
Sánchez-Calderón, L., Chacón-López, A., Alatorre-Cobos, F., Leyva-González, M. A. & Herrera-Estrella, L. in Transporters and Pumps in Plant Signaling (eds Markus Geisler & Kees Venema) 191–224 (Springer, 2011).
Poitout, A. et al. Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-Zeatin in shoots. Plant Cell 30, 1243–1257 (2018).
Takei, K. et al. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45, 1053–1062 (2004).
Martin, A. C. et al. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 24, 559–567 (2000).
Wang, X. M. et al. Cytokinin represses phosphate-starvation response through increasing of intracellular phosphate level. Plant Cell Environ. 29, 1924–1935 (2006).
Lu, Y. et al. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 10, 1242–1245 (2017).
Hellens, R. P. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005).
Chen, H. et al. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 146, 368–376 (2008).
Luo, A. et al. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47, 181–191 (2006).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence-analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994).
Wang, C. et al. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 57, 895–904 (2009).
Hu, B. et al. Leaf tip necrosis1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 156, 1101–1115 (2011).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1672 (2007).
Zhang, Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30 (2011).
Hecker, A. et al. Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiol. 168, 776–787 (2015).
Zhao, Q., Liu, L. & Xie, Q. In vitro protein ubiquitination assay. Methods Mol. Biol. 876, 163–172 (2012).
Zhao, Q. et al. A plant‐specific in vitro ubiquitination analysis system. Plant J. 74, 524–533 (2013).
Wang, Z. et al. SCFSAP controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nat. Commun. 7, 11192 (2016).
van Dongen, W., Van Heerde, L., Boeren, S. & De Vries, S. C. Identification of brassinosteroid signaling complexes by coimmunoprecipitation and mass spectrometry. Methods Mol. Biol. 1564, 145–154 (2017).
Acknowledgements
pCAMBIA1300–nLUC/cLUC vectors were kindly provided by J.-M. Zhou, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. The SPX4pro–gSPX4–GUS transgenic rice was kindly provided by C. Mao, College of Life Sciences, Zhejiang University. The authors thank T. O. Jobe (University of Cologne) for proofreading the manuscript. This work was supported by grants from the Chinese Academy of Sciences (XDA08010400), the National Natural Sciences Foundation of China (31500975 and 31771348) and the China Postdoctoral Science Foundation (2017M610126). Research in the laboratory of S. Kopriva was supported by Deutsche Forschungsgemeinschaft (EXC 1028).
Author information
Authors and Affiliations
Contributions
B.H., Z.J. and W.W. designed research, performed experiments, analysed the data and wrote the manuscript. Y. Qiu, Z.Z., Y.L., A.L., X.G., L.L., Y. Qian, X.H., F.Y., S. Kang, Yiq. W., J.X., S.C., L.Z., Ying. W., Q.X. and S. Kopriva conducted some of the experiments. C.C. designed research, wrote the manuscript and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Plants thanks Dong Liu, Nicholaus von Wiren, Ying Liu, Ricardo Giehl and other anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–14 and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Hu, B., Jiang, Z., Wang, W. et al. Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413 (2019). https://doi.org/10.1038/s41477-019-0384-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0384-1
This article is cited by
-
Role of GARP family transcription factors in the regulatory network for nitrogen and phosphorus acquisition
Journal of Plant Research (2024)
-
Strigolactone regulates nitrogen-phosphorus balance in rice
Science China Life Sciences (2024)
-
Reviewing impacts of biotic and abiotic stresses on the regulation of phosphate homeostasis in plants
Journal of Plant Research (2024)
-
Synergistic effects of leaf nitrogen and phosphorus on photosynthetic capacity in subtropical forest
Theoretical and Experimental Plant Physiology (2024)
-
OsCBL1 modulates rice nitrogen use efficiency via negative regulation of OsNRT2.2 by OsCCA1
BMC Plant Biology (2023)