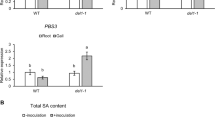
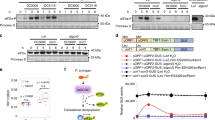
Abstract
Striga parasitizes major crops in arid regions, depriving the host crop of nutrients through the transpiration stream and causing vast agricultural damage. Here, we report on the mechanism underlying how Striga maintains high transpiration under drought conditions. We found that Striga did not respond to abscisic acid, the phytohormone responsible for controlling stomatal closure. Protein phosphatase 2C of Striga (ShPP2C1) is not regulated by abscisic acid receptors, and this feature is attributable to specific mutations in its amino acid sequence. Moreover, Arabidopsis transformed with ShPP2C1 showed an abscisic acid-insensitive phenotype, indicating that ShPP2C1 functions as a dominant negative regulator of abscisic acid signal transduction. These findings suggest that ShPP2C1 interrupts abscisic acid signalling in Striga, resulting in high transpiration and subsequent efficient absorption of host nutrients under drought conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The RNA sequencing data have been deposited at the DNA Databank of Japan under accession codes DRX142184, DRX142185. Complementary DNAs were registered at the DNA Databank of Japan with accession numbers as follows: ShPYL1, LC377366; ShPYL2, LC377367; ShPYL3, LC377368; ShPYL4, LC377369; ShPYL5, LC377370; ShPYL6, LC377371; ShPYL7, LC377372; ShPYL8, LC377373; ShPP2C1, LC377362; ShPP2C2, LC377363; ShPP2C3, LC377364; ShPP2C4, LC377365. Other data that support the findings of this study are available from the corresponding author on reasonable request.
References
Gobena, D. et al. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc. Natl Acad. Sci. USA 114, 4471–4476 (2017).
Graves, J. D. et al. A carbon balance model of the sorghum–Striga hermonthica host–parasite association. Plant Cell Environ. 12, 101–107 (1989).
Dörr, I. How Striga parastizes its host: a TEM and SEM study. Ann. Bot. 79, 463–472 (1997).
Ejeta, G. & Gressel, J. (eds) Integrating new technologies for Striga control. 27–28 (World Scientific, 2007).
Ackroyd, R. D. & Graves, J. D. The regulation of the water potential gradient in the host and parasite relationship between Sorghum bicolor and Striga hermonthica. Ann. Bot. 80, 649–656 (1997).
Inoue, T. et al. Gas exchange of root hemi-parasite Striga hermonthica and its host Sorghum bicolor under short-term soil water stress. Biol. Plant. 57, 773–777 (2013).
Finkelstein, R. Abscisic acid synthesis. Arab. B. 11, e0166 (2013).
Taylor, A. et al. Physiology of the parasitic association between maize and witchweed (Striga hermonthica): is ABA involved? J. Exp. Bot. 47, 1057–1065 (1996).
Zwanenburg, B. et al. Structure and function of natural and synthetic signalling molecules in parasitic weed germination. Pest Manag. Sci. 65, 478–491 (2009).
Pouvreau, J. B. et al. A high-throughput seed germination assay for root parasitic plants. Plant Methods 9, 32 (2013).
Cutler, S. R. et al. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679 (2010).
Koornneef, M. et al. The isolation and characterization of abscisic acid‐insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61, 377–383 (1984).
Leung, J. et al. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264, 1448–1452 (1994).
Leung, J. et al. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–71 (1997).
Fujii, H. et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664 (2010).
Mosquna, A. et al. Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc. Natl Acad. Sci. USA 108, 20838–20843 (2011).
Toh, S. et al. Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol. 53, 107–117 (2012).
Miyazono, K. et al. Structural basis of abscisic acid signalling. Nature 462, 609–614 (2009).
Dupeux, F. et al. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol. 156, 106–116 (2011).
Melcher, K. et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608 (2009).
Bowman, J. L. et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304 (2017).
Aly, R. et al. Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol. J. 7, 487–498 (2009).
Delavault, P. et al. Isolation of mannose 6-phosphate reductase cDNA, changes in enzyme activity and mannitol content in broomrape. Physiol. Plant. 115, 48–55 (2002).
Acknowledgements
We thank A.G.T. Babiker for providing Striga seeds. We also thank T. Hosouchi and S. Shinpo for technical support in Illumina sequencing; N. Arimoto, M. Negishi and Y. Yamakawa for supporting transgenic Arabidopsis experiments; Y. Tsuchiya and K. Hanada for advising on identification of the ABA receptor and PP2C from Striga; and T. Hirayama for providing the Columbia accession background Arabidopsis abi1c mutant. This work was supported by grants from MEXT/JSPS KAKENHI (No. 17H05009 to M.O., No. 15H05248 to Y.S.), MEXT/JSPS Bilateral Programs (to Y.S.), the Joint Research Program of Arid Land Research Center, Tottori University (No. 28C2004 to Y.S.) and Japan Science and Technology Agency (PRESTO, No. JPMJPR15Q5 to M.O.); and the SATREPS Striga project to Y.S.
Author information
Authors and Affiliations
Contributions
Y.S. and M.O. conceived and designed the project. H.F. and H.S. contributed to physiological research on S. hermonthica. H.F. performed gene expression analysis, protein expression, purification and biochemical assays. H.S. and M.M. performed next generation sequencing and data analysis. M.O. carried out molecular cloning, transgenic plant generation and physiological analysis of transformants. H.F., M.O. and Y.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–8, Supplementary Table 1, Supplementary Methods and Supplementary References.
Rights and permissions
About this article
Cite this article
Fujioka, H., Samejima, H., Suzuki, H. et al. Aberrant protein phosphatase 2C leads to abscisic acid insensitivity and high transpiration in parasitic Striga. Nat. Plants 5, 258–262 (2019). https://doi.org/10.1038/s41477-019-0362-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0362-7
This article is cited by
-
Nitrogen represses haustoria formation through abscisic acid in the parasitic plant Phtheirospermum japonicum
Nature Communications (2022)
-
ABA signaling components in Phelipanche aegyptiaca
Scientific Reports (2019)