Abstract
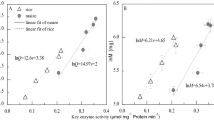
Plants modulate their growth rate according to seasonal and environmental cues using a suite of growth repressors known to interact directly with cellular machinery controlling cell division and growth. Mutants lacking growth repressors show increased growth rates1,2, but the mechanism by which these plants ensure source availability for faster growth is unclear. Here, we undertake a comprehensive analysis of the fast-growth phenotype of a quintuple growth-repressor mutant, using a combination of theoretical and experimental approaches to understand the physiological basis of source–sink coordination. Our results show that, in addition to the control of tissue growth rates, growth repressors also affect tissue composition and leaf thickness, modulating the efficiency of production of new photosynthetic capacity. Modelling suggests that increases in growth efficiency underlie growth-rate differences between the wild type and spatula della growth-repressor mutant, with spatula della requiring less carbon to synthesize a comparable photosynthetic capability to the wild type, and fixing more carbon per unit mass. We conclude that through control of leaf development, growth repressors regulate both source availability and sink strength to achieve growth-rate variation without risking a carbon deficit.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data generated and analysed during this study are available from the corresponding author on reasonable request.
References
Sidaway-Lee, K. et al. SPATULA links daytime temperature and plant growth rate. Curr. Biol. 20, 1493–1497 (2010).
Achard, P. et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (2006).
Sitch, S. et al. Evaluation of ecosystem dynamics, plant geography and terrestrial carbon cycling in the LPJ dynamic global vegetation model. Glob. Change Biol. 9, 161–185 (2003).
Peng, J. et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205 (1997).
Silverstone, A. L. et al. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1566 (2001).
Koini, M. A. et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413 (2009).
Ichihashi, Y., Horiguchi, G., Gleissberg, S. & Tsukaya, H. The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 51, 252–261 (2010).
Serrano-Mislata, A. et al. DELLA genes restrict inflorescence meristem function independently of plant height. Nat. Plants 3, 749–754 (2017).
White, A. C., Rogers, A., Rees, M. & Osborne, C. P. How can we make plants grow faster? A source–sink perspective on growth rate. J. Exp. Bot. 67, 31–45 (2015).
Körner, C. Paradigm shift in plant growth control. Curr. Opin. Plant Biol. 25, 107–114 (2015).
Josse, E. M. et al. A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. Plant Cell 23, 1337–1351 (2011).
Achard, P. et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129 (2008).
Makkena, S. & Lamb, R. S. The bHLH transcription factor SPATULA regulates root growth by controlling the size of the root meristem. BMC Plant Biol. 13, 1 (2013).
Ribeiro, D. M., Araújo, W. L., Fernie, A. R., Schippers, J. H. & Mueller-Roeber, B. Action of gibberellins on growth and metabolism of Arabidopsis plants associated with high concentration of carbon dioxide. Plant Physiol. 160, 1781–1794 (2012).
Paparelli, E. et al. Nighttime sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis. Plant Cell 25, 3760–3769 (2013).
Achard, P. et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193 (2009).
Tyler, L. et al. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019 (2004).
Penfield, S. et al. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15, 1998–2006 (2005).
Barth, S., Busimi, A., Utz, H. F. & Melchinger, A. Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity 91, 36–42 (2003).
Meyer, R. C., Törjék, O., Becher, M. & Altmann, T. Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 134, 1813–1823 (2004).
Boyes, D. C. et al. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510 (2001).
Chew, Y. H. et al. Multiscale digital Arabidopsis predicts individual organ and whole-organism growth. Proc. Natl Acad. Sci. USA 111, E4127–E4136 (2014).
Rasse, D. P. & Tocquin, P. Leaf carbohydrate controls over Arabidopsis growth and response to elevated CO2: an experimentally based model. New Phytol. 172, 500–513 (2006).
Christophe, A. et al. A model-based analysis of the dynamics of carbon balance at the whole-plant level in Arabidopsis thaliana. Funct. Plant Biol. 35, 1147–1162 (2008).
Zhou, J. et al. Leaf-GP: an open and automated software application for measuring growth phenotypes for Arabidopsis and wheat. Plant Methods 13, 117 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Friedland, G., Jantz, K. & Rojas, R. SIOX: simple interactive object extraction in still images. In 7th IEEE International Symposium on Multimedia 7–13 (IEEE, 2005).
Conn, S. J. et al. Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9, 4 (2013).
R Development Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, New York, 2016).
Eaton, J. W., Bateman, D., Hauberg, S. & Wehbring, R. GNU Octave Version 3.8.1 Manual: A High-Level Interactive Language for Numerical Computations (CreateSpace Independent Publishing Platform, 2014).
Acknowledgements
This work was supported by a Leverhulme Trust Research Project Grant.
Author information
Authors and Affiliations
Contributions
N.P. and S.P. designed the experiments. N.Z., A.D.A. and N.P. performed the experiments. N.P. analysed the data and performed the modelling. N.P. and S.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–5, Supplementary Methods and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Pullen, N., Zhang, N., Dobon Alonso, A. et al. Growth rate regulation is associated with developmental modification of source efficiency. Nature Plants 5, 148–152 (2019). https://doi.org/10.1038/s41477-018-0357-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0357-9