Abstract
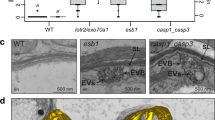
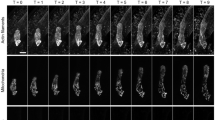
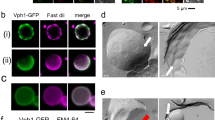
Plant vacuoles are dynamic organelles that play essential roles in regulating growth and development. Two distinct models of vacuole biogenesis have been proposed: separate vacuoles are formed by the fusion of endosomes, or the single interconnected vacuole is derived from the endoplasmic reticulum. These two models are based on studies of two-dimensional (2D) transmission electron microscopy and 3D confocal imaging, respectively. Here, we performed 3D electron tomography at nanometre resolution to illustrate vacuole biogenesis in Arabidopsis root cells. The whole-cell electron tomography analysis first identified unique small vacuoles (SVs; 400–1,000 nm in diameter) as nascent vacuoles in early developmental cortical cells. These SVs contained intraluminal vesicles and were mainly derived/matured from multivesicular body (MVB) fusion. The whole-cell vacuole models and statistical analysis on wild-type root cells of different vacuole developmental stages demonstrated that central vacuoles were derived from MVB-to-SV transition and subsequent fusions of SVs. Further electron tomography analysis on mutants defective in MVB formation/maturation or vacuole fusion demonstrated that central vacuole formation required functional MVBs and membrane fusion machineries.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
References
Saftig, P. & Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 (2009).
Luzio, J. P., Hackmann, Y., Dieckmann, N. M. & Griffiths, G. M. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb. Perspect. Biol. 6, a016840 (2014).
Armstrong, J. Yeast vacuoles: more than a model lysosome. Trends Cell Biol. 20, 580–585 (2010).
Bryant, N. J. & Stevens, T. H. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62, 230–247 (1998).
Shimada, T., Takagi, J., Ichino, T., Shirakawa, M. & Hara-Nishimura, I. Plant vacuoles. Annu. Rev. Plant Biol. 69, 123–145 (2018).
Paris, N., Stanley, C. M., Jones, R. L. & Rogers, J. C. Plant cells contain two functionally distinct vacuolar compartments. Cell 85, 563–572 (1996).
Feeney, M., Kittelmann, M., Menassa, R., Hawes, C. & Frigerio, L. Protein storage vacuoles originate from remodeled preexisting vacuoles in Arabidopsis thaliana. Plant Physiol. 177, 241–254 (2018).
Zheng, H. Q. & Staehelin, L. A. Protein storage vacuoles are transformed into lytic vacuoles in root meristematic cells of germinating seedlings by multiple, cell type-specific mechanisms. Plant Physiol. 155, 2023–2035 (2011).
Scheres, B. & Wolkenfelt, H. The Arabidopsis root as a model to study plant development. Plant Physiol. Biochem. 36, 21–32 (1998).
Scheres, B., Benfey, P. & Dolan, L. Root development. Arabidopsis Book 1, e0101 (2002).
Zouhar, J. & Rojo, E. Plant vacuoles: where did they come from and where are they heading? Curr. Opin. Plant Biol. 12, 677–684 (2009).
Eisenach, C., Francisco, R. & Martinoia, E. Plant vacuoles. Curr. Biol. 25, R136–R137 (2015).
Zhang, C. H., Hicks, G. R. & Raikhel, N. V. Plant vacuole morphology and vacuolar trafficking. Front. Plant Sci. 5, 476 (2014).
Viotti, C. ER and vacuoles: never been closer. Front. Plant Sci. 5, 20 (2014).
Marty, F. Plant vacuoles. Plant Cell 11, 587–599 (1999).
Marty, F. Cytochemical studies on Gerl, provacuoles, and vacuoles in root meristematic cells of Euphorbia. Proc. Natl Acad. Sci. USA 75, 852–856 (1978).
Viotti, C. et al. The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25, 3434–3449 (2013).
Amelunxen, F. & Heinze, U. On the development of the vacuole in the testa cells of linum seeds. Eur. J. Cell Biol. 35, 343–354 (1984).
Scheuring, D. et al. Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. Proc. Natl Acad. Sci. USA 113, 452–457 (2016).
Lofke, C., Dunser, K., Scheuring, D. & Kleine-Vehn, J. Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. eLife 4, e05868 (2015).
Kolb, C. et al. FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 167, 1361–1373 (2015).
Kalinowska, K. et al. Arabidopsis ALIX is required for the endosomal localization of the deubiquitinating enzyme AMSH3. Proc. Natl Acad. Sci. USA 112, E5543–E5551 (2015).
Wang, J., Cai, Y., Miao, Y., Lam, S. K. & Jiang, L. Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60, 3075–3083 (2009).
Tse, Y. C. et al. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16, 672–693 (2004).
Miao, Y. & Jiang, L. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2, 2348–2353 (2007).
Scheuring, D. et al. Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23, 3463–3481 (2011).
Buono, R. A. et al. ESCRT-mediated vesicle concatenation in plant endosomes. J. Cell Biol. 216, 2167–2177 (2017).
Takano, J., Miwa, K., Yuan, L. X., von Wiren, N. & Fujiwara, T. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl Acad. Sci. USA 102, 12276–12281 (2005).
Zheng, J. M., Han, S. W., Rodriguez-Welsh, M. F. & Rojas-Pierce, M. Homotypic vacuole fusion requires VTI11 and is regulated by phosphoinositides. Mol. Plant 7, 1026–1040 (2014).
Takemoto, K. et al. Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl Acad. Sci. USA 115, E2457–E2466 (2018).
Geldner, N. et al. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178 (2009).
Gao, C., Zhuang, X., Shen, J. & Jiang, L. Plant ESCRT complexes: moving beyond endosomal sorting. Trends Plant Sci. 22, 986–998 (2017).
Gao, C. et al. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 24, 2556–2563 (2014).
Singh, M. K. et al. Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB–vacuole fusion. Curr. Biol. 24, 1383–1389 (2014).
Ebine, K. et al. Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr. Biol. 24, 1375–1382 (2014).
Cui, Y. et al. Activation of the Rab7 GTPase by the MON1–CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26, 2080–2097 (2014).
Segui-Simarro, J. M. & Staehelin, L. A. Cell cycle-dependent changes in Golgi stacks, vacuoles, clathrin-coated vesicles and multivesicular bodies in meristematic cells of Arabidopsis thaliana: a quantitative and spatial analysis. Planta 223, 223–236 (2006).
Zhuang, X. et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl Acad. Sci. USA 114, E426–E435 (2017).
Ivanov, R. & Robinson, D. G. Turnover of tonoplast proteins. Plant Physiol. 177, 10–11 (2018).
Maitrejean, M. & Vitale, A. How are tonoplast proteins degraded? Plant Signal. Behav. 6, 1809–1812 (2011).
Cui, Y. et al. Biogenesis of plant prevacuolar multivesicular bodies. Mol. Plant 9, 774–786 (2016).
Jauh, G. Y., Phillips, T. E. & Rogers, J. C. Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell 11, 1867–1882 (1999).
Zwiewka, M. et al. The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21, 1711–1722 (2011).
Bottanelli, F., Foresti, O., Hanton, S. & Denecke, J. Vacuolar transport in tobacco leaf epidermis cells involves a single route for soluble cargo and multiple routes for membrane cargo. Plant Cell 23, 3007–3025 (2011).
Uemura, T. & Ueda, T. Plant vacuolar trafficking driven by Rab and SNARE proteins. Curr. Opin. Plant Biol. 22, 116–121 (2014).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Cui, Y. et al. MONENSIN SENSITIVITY1 (MON1)/CALCIUM CAFFEINE ZINC SENSITIVITY1 (CCZ1)-mediated Rab7 activation regulates tapetal programmed cell death and pollen development. Plant Physiol. 173, 206–218 (2017).
Kang, B. H. Electron microscopy and high-pressure freezing of Arabidopsis. Method Cell Biol. 96, 259–283 (2010).
Sattarzadeh, A., Saberianfar, R., Zipfel, W. R., Menassa, R. & Hanson, M. R. Green to red photoconversion of GFP for protein tracking in vivo. Sci. Rep. 5, 11771 (2015).
Acknowledgements
This work was supported by grants from the Research Grants Council of Hong Kong (CUHK14130716, 14102417, 14100818, C4011-14R, C4012-16E, C4002-17G and AoE/M-05/12) and the National Natural Science Foundation of China (31270226, 31470294 and 91854201).
Author information
Authors and Affiliations
Contributions
Y.C., B.-H.K. and L.J. conceived and designed the experiments. Y.C. performed the electron tomography analysis. Y.C., W.C., Y.H., H.Y.W., W.S.W. and H.K.L. generated the 3D models. Y.C., W.C., Y.H., Q.Z., M.W., X.Z., J.G., Y.Z., C.G., Y.D. and P.W. performed the other experiments. Y.C., T.U., M.R.-P., K.T., B.-H.K. and L.J. analysed the data. Y.C. and L.J. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–8.
Supplementary Video 1
Whole-cell electron tomography analyses of SVs in relationship with other organelles in Cell 1.
Supplementary Video 2
3D tomography analyses of detailed structures and relationships of ER, Golgi, TGN, MVBs and SVs.
Supplementary Video 3
3D tomography analyses of fusion between MVBs and SVs, along with a transfer of ILVs.
Supplementary Video 4
Whole-cell electron tomography analyses of vacuoles in Cell 2.
Supplementary Video 5
Whole-cell electron tomography analyses of vacuoles in Cell 3.
Supplementary Video 6
3D tomography analyses of vacuoles in WT and various mutants.
Rights and permissions
About this article
Cite this article
Cui, Y., Cao, W., He, Y. et al. A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nature Plants 5, 95–105 (2019). https://doi.org/10.1038/s41477-018-0328-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0328-1
This article is cited by
-
Vacuolar control of stomatal opening revealed by 3D imaging of the guard cells
Scientific Reports (2023)
-
Cellular dynamics of coenocytic endosperm development in Arabidopsis thaliana
Nature Plants (2023)
-
The plant unique ESCRT component FREE1 regulates autophagosome closure
Nature Communications (2023)
-
Plantorganelle Hunter is an effective deep-learning-based method for plant organelle phenotyping in electron microscopy
Nature Plants (2023)
-
Plant-derived extracellular vesicles: a novel nanomedicine approach with advantages and challenges
Cell Communication and Signaling (2022)