Abstract
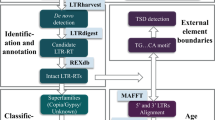
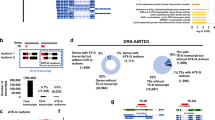
Retrotransposons have played an important role in the evolution of host genomes1,2. Their impact is mainly deduced from the composition of DNA sequences that have been fixed over evolutionary time2. Such studies provide important ‘snapshots’ reflecting the historical activities of transposons but do not predict current transposition potential. We previously reported sequence-independent retrotransposon trapping (SIRT) as a method that, by identification of extrachromosomal linear DNA (eclDNA), revealed the presence of active long terminal repeat (LTR) retrotransposons in Arabidopsis3. However, SIRT cannot be applied to large and transposon-rich genomes, as found in crop plants. We have developed an alternative approach named ALE-seq (amplification of LTR of eclDNAs followed by sequencing) for such situations. ALE-seq reveals sequences of 5′ LTRs of eclDNAs after two-step amplification: in vitro transcription and subsequent reverse transcription. Using ALE-seq in rice, we detected eclDNAs for a novel Copia family LTR retrotransposon, Go-on, which is activated by heat stress. Sequencing of rice accessions revealed that Go-on has preferentially accumulated in Oryza sativa ssp. indica rice grown at higher temperatures. Furthermore, ALE-seq applied to tomato fruits identified a developmentally regulated Gypsy family of retrotransposons. A bioinformatic pipeline adapted for ALE-seq data analyses is used for the direct and reference-free annotation of new, active retroelements. This pipeline allows assessment of LTR retrotransposon activities in organisms for which genomic sequences and/or reference genomes are either unavailable or of low quality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The next-generation sequencing data that support the findings of this study are available in the Sequence Read Archive (SRA) repository with the identifier SRP155920.
References
Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 14, 49–61 (2012).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017).
Griffiths, J., Catoni, M., Iwasaki, M. & Paszkowski, J. Sequence-independent identification of active LTR retrotransposons in Arabidopsis. Mol. Plant 11, 508–511 (2017).
Ma, J. & Bennetzen, J. L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl Acad. Sci. USA 101, 12404–12410 (2004).
Sanchez, D. H., Gaubert, H., Drost, H., Zabet, N. R. & Paszkowski, J. High-frequency recombination between members of an LTR retrotransposon family during transposition bursts. Nat. Commun. 8, 1283 (2017).
Picault, N. et al. Identification of an active LTR retrotransposon in rice. Plant J. 58, 754–765 (2009).
Sabot, F. et al. Transpositional landscape of the rice genome revealed by paired-end mapping of high-throughput re-sequencing data. Plant J. 66, 241–246 (2011).
Mirouze, M. et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461, 427–430 (2009).
Ito, H. et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119 (2011).
Paszkowski, J. Controlled activation of retrotransposition for plant breeding. Curr. Opin. Biotechnol. 32, 200–206 (2015).
Cavrak, V. V. et al. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 10, e1004115 (2014).
Pietzenuk, B. et al. Recurrent evolution of heat-responsiveness in Brassicaceae COPIA elements. Genome Biol. 17, 209 (2016).
3K RGP. The 3,000 rice genomes project. Gigascience 3, 7 (2014).
Nakagome, M. et al. Transposon insertion finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinform. 15, 71 (2014).
Xiong, Z. Y. et al. Latitudinal distribution and differentiation of rice germplasm: its implications in breeding. Crop Sci. 51, 1050–1058 (2011).
Zhong, S. et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159 (2013).
The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Eshed, Y. & Zamir, D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995).
Eshed, Y. & Zamir, D. Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 143, 1807–1817 (1996).
Quadrana, L. et al. The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5, e15716 (2016).
Stuart, T. et al. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife 5, e20777 (2016).
Wei, B. et al. Genome-wide characterization of non-reference transposons in crops suggests non-random insertion. BMC Genomics 17, 536 (2016).
Lanciano, S. et al. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet. 13, e1006630 (2017).
Møller, H. D. et al. Formation of extrachromosomal circular DNA from long terminal repeats of retrotransposons in Saccharomyces cerevisiae. G3 (Bethesda) 6, 453–462 (2015).
Møller, H. D., Parsons, L., Jørgensen, T. S., Botstein, D. & Regenberg, B. Extrachromosomal circular DNA is common in yeast. Proc. Natl Acad. Sci. USA 112, 3114–3122 (2015).
Cheng, C. et al. Loss of function mutations in the rice chromomethylase OsCMT3a cause a burst of transposition. Plant J. 83, 1069–1081 (2015).
Cui, X. et al. Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl Acad. Sci. USA 110, 1953–1958 (2013).
Wang, Z. H. et al. Genomewide variation in an introgression line of rice-zizania revealed by whole-genome re-sequencing. PLoS ONE 8, e74479 (2013).
Li, H., Freeling, M. & Lisch, D. Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl Acad. Sci. USA 107, 22184–22189 (2010).
Slotkin, R. K. et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472 (2009).
Liu, R. et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl Acad. Sci. USA 112, 10804–10809 (2015).
Goodier, J. L. Retrotransposition in tumors and brains. Mob. DNA 5, 11 (2014).
Baillie, J. K. et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537 (2011).
Mullins, C. S. & Linnebacher, M. Human endogenous retroviruses and cancer: causality and therapeutic possibilities. World J. Gastroenterol. 18, 6027–6035 (2012).
Cho, J. & Paszkowski, J. Regulation of rice root development by a retrotransposon acting as a microRNA sponge. eLife 6, e30038 (2017).
Chan, P. P. & Lowe, T. M. GtRNAdb 2. 0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, 184–189 (2016).
Daujat, M. et al. PlantRNA, a database for tRNAs of photosynthetic eukaryotes. Nucleic Acids Res. 41, 273–279 (2012).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, 222–230 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 516–520 (2010).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Catoni, M. et al. DNA sequence properties that predict susceptibility to epiallelic switching. EMBO J. 36, 617–628 (2017).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011).
Catoni, M., Tsang, J. M. F., Greco, A. P. & Zabet, N. R. DMRcaller: a versatile R/Bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Res. 46, e114 (2018).
Lawrence, M., Gentleman, R. & Carey, V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25, 1841–1842 (2009).
Acknowledgements
This work was supported by European Research Council (EVOBREED) grant no. 322621 and a Gatsby Fellowship grant no. AT3273/GLE.
Author information
Authors and Affiliations
Contributions
J.C. and J.P. conceived the research. J.C., M.B., M.C., H.-G.D., A.B. and M.O. performed experiments. J.C., M.B., M.C. and H.-G.D. analysed data. J.C. and J.P. wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–11 and Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Cho, J., Benoit, M., Catoni, M. et al. Sensitive detection of pre-integration intermediates of long terminal repeat retrotransposons in crop plants. Nature Plants 5, 26–33 (2019). https://doi.org/10.1038/s41477-018-0320-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0320-9
This article is cited by
-
Transposable element-initiated enhancer-like elements generate the subgenome-biased spike specificity of polyploid wheat
Nature Communications (2023)
-
Amplification of LTRs of extrachromosomal linear DNAs (ALE-seq) identifies two active Oryco LTR retrotransposons in the rice cultivar Dongjin
Mobile DNA (2022)
-
Harnessing epigenetic variability for crop improvement: current status and future prospects
Genes & Genomics (2022)
-
Pararetroviruses: Plant Infecting dsDNA Viruses
Plant Molecular Biology Reporter (2022)
-
Recent advancement of NGS technologies to detect active transposable elements in plants
Genes & Genomics (2021)