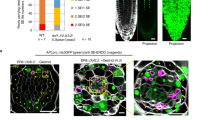
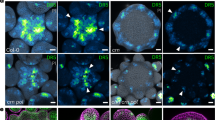
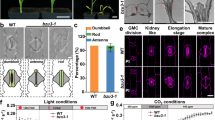
Abstract
The frequency and orientation of cell division are regulated by intercellular signalling molecules; however, tissue-specific regulatory systems for cell divisions are only partially understood. Here, we report that the peptide hormone CLAVATA3/ESR-RELATED 9/10 (CLE9/10) regulates two different developmental processes, stomatal lineage development and xylem development, through two distinct receptor systems in Arabidopsis thaliana. We show that the receptor kinase HAESA-LIKE 1 (HSL1) is a CLE9/10 receptor that regulates stomatal lineage cell division, and BARELY NO MERISTEM (BAM) class receptor kinases are CLE9/10 receptors that regulate periclinal cell division of xylem precursor cells. Both HSL1 and BAM1 bind to CLE9/10, but only HSL1 recruits SOMATIC EMBRYOGENESIS RECEPTOR KINASES as co-receptors in the presence of CLE9/10, suggesting different signalling modes for these receptor systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Change history
26 December 2018
In the version of this Article originally published, the authors incorrectly stated that the work was supported by Innovative Areas grant number 25003006; the correct number is 25113006. This statement has now been amended in all online versions of the Article.
References
Yamaguchi, Y. L., Ishida, T. & Sawa, S. CLE peptides and their signaling pathways in plant development. J. Exp. Bot. 67, 4813–4826 (2016).
Fletcher, J. C., Brand, U., Running, M. P., Simon, R. & Meyerowitz, E. M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914 (1999).
Shinohara, H. & Matsubayashi, Y. Reevaluation of the CLV3–receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 82, 328–336 (2015).
Hirakawa, Y., Kondo, Y. & Fukuda, H. Establishment and maintenance of vascular cell communities through local signaling. Curr. Opin. Plant Biol. 14, 17–23 (2011).
Kondo, Y., Hirakawa, Y., Kieber, J. J. & Fukuda, H. CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 52, 37–48 (2011).
Pillitteri, L. J. & Torii, K. U. Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63, 591–614 (2012).
Geisler, M., Nadeau, J. & Sack, F. D. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12, 2075–2086 (2000).
Bergmann, D. C. & Sack, F. D. Stomatal development. Annu. Rev. Plant Biol. 58, 163–181 (2007).
Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L. & Torii, K. U. Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 (2007).
MacAlister, C. A., Ohashi-Ito, K. & Bergmann, D. C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 (2007).
Lampard, G. R., MacAlister, C. A. & Bergmann, D. C. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116 (2008).
Meng, X. et al. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361–2372 (2015).
Lee, J. S. et al. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522, 439–443 (2015).
Jewaria, P. K. et al. Differential effects of the peptides Stomagen, EPF1 and EPF2 on activation of MAP kinase MPK6 and the SPCH protein level. Plant Cell Physiol. 54, 1253–1262 (2013).
Hara, K. et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031 (2009).
De Rybel, B., Mahonen, A. P., Helariutta, Y. & Weijers, D. Plant vascular development: from early specification to differentiation. Nat. Rev. Mol. Cell Biol. 17, 30–40 (2016).
Bergmann, D. C., Lukowitz, W. & Somerville, C. R. Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497 (2004).
Pillitteri, L. J., Peterson, K. M., Horst, R. J. & Torii, K. U. Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23, 3260–3275 (2011).
Adrian, J. et al. Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev. Cell 33, 107–118 (2015).
Jun, J. et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154, 1721–1736 (2010).
Vaten, A., Soyars, C. L., Tarr, P. T., Nimchuk, Z. L. & Bergmann, D. C. Modulation of asymmetric division diversity through cytokinin and SPEECHLESS regulatory interactions in the Arabidopsis stomatal lineage. Dev. Cell 47, 53–66 (2018).
Shinohara, H., Moriyama, Y., Ohyama, K. & Matsubayashi, Y. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 70, 845–854 (2012).
Rodriguez-Villalon, A., Gujas, B., van Wijk, R., Munnik, T. & Hardtke, C. S. Primary root protophloem differentiation requires balanced phosphatidylinositol-4,5-biphosphate levels and systemically affects root branching. Development 142, 1437–1446 (2015).
Kang, Y. H. & Hardtke, C. S. Arabidopsis MAKR5 is a positive effector of BAM3-dependent CLE45 signaling. EMBO Rep. 17, 1145–1154 (2016).
Hazak, O. & Hardtke, C. S. CLAVATA 1-type receptors in plant development. J. Exp. Bot. 67, 4827–4833 (2016).
Hazak, O. et al. Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. EMBO Rep. 18, 1367–1381 (2017).
Czyzewicz, N. et al. Antagonistic peptide technology for functional dissection of CLE peptides revisited. J. Exp. Bot. 66, 5367–5374 (2015).
Anne, P. et al. CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development 145, dev162354 (2018).
Shiu, S. H. & Bleecker, A. B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. USA 98, 10763–10768 (2001).
Song, W., Han, Z., Wang, J., Lin, G. & Chai, J. Structural insights into ligand recognition and activation of plant receptor kinases. Curr. Opin. Struct. Biol. 43, 18–27 (2016).
Matsubayashi, Y. Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413 (2014).
Song, W. et al. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26, 674–685 (2016).
Santiago, J. et al. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5, e15075 (2016).
Ma, X., Xu, G., He, P. & Shan, L. SERKing coreceptors for receptors. Trends Plant Sci. 21, 1017–1033 (2016).
Zhang, H., Lin, X., Han, Z., Qu, L. J. & Chai, J. Crystal structure of PXY–TDIF complex reveals a conserved recognition mechanism among CLE peptide–receptor pairs. Cell Res. 26, 543–555 (2016).
Carlsbecker, A. et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010).
Kumari, A., Jewaria, P. K., Bergmann, D. C. & Kakimoto, T. Arabidopsis reduces growth under osmotic stress by decreasing SPEECHLESS protein. Plant Cell Physiol. 55, 2037–2046 (2014).
Nimchuk, Z. L., Tarr, P. T., Ohno, C., Qu, X. & Meyerowitz, E. M. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21, 345–352 (2011).
Hu, C. et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4, 205–211 (2018).
Alberts, B. J. A. et al. Molecular Biology of the Cell (Garland Science, New York, 2014).
Gordon, M. D. & Nusse, R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 281, 22429–22433 (2006).
Shpak, E. D., McAbee, J. M., Pillitteri, L. J. & Torii, K. U. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293 (2005).
Hara, K., Kajita, R., Torii, K. U., Bergmann, D. C. & Kakimoto, T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720–1725 (2007).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Kurihara, D., Mizuta, Y., Sato, Y. & Higashiyama, T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015).
Wang, Z. P. et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015).
Motohashi, K. A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol. 15, 47 (2015).
Pedelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Shaner, N. C. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013).
Acknowledgements
Special thanks to H. Fukuda and Y. Kondo for providing us with all of the synthetic CLE peptides, Q.-J. Chen for the CRISPR vector (pHEE401E), K. Torii, D. Bergmann, S. Hou and the Arabidopsis Biological Resource Center seed stock centre for mutant seeds, and H. Deng and Y. Xu for mass spectrometry analysis. We thank K. Tsujimura for making vectors containing histone H2BsfGFP and mNEONGREEN optimized for A. thaliana. We thank Y. Matsubayashi and H. Shinohara for providing us with bam1-4 and bam1-4 clv1-101 mutants, discussions and preliminary experiments, and B. Morris and A. Deneve for advice on English usage. This work was supported by Grant-in-Aid for Scientific Research (B) number 25291060, Innovative Areas grant numbers 25113006 and 18H04837 to T.K. and the Ministry of Science and Technology of China (2015CB910200) to J.C.
Author information
Authors and Affiliations
Contributions
P.Q., W.S., T.Y., A.M., G.W. and T.I. performed the experiments. P.Q., T.I., S.S., J.C. and T.K. designed the research. P.Q., W.S., J.C. and T.K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–21 and Supplementary Tables 1–3.
Supplementary Video 1
Periclinal cell divisions of xylem precursor cells in wild type.
Supplementary Video 2
Periclinal cell divisions of xylem precursor cells in cle9-c1.
Rights and permissions
About this article
Cite this article
Qian, P., Song, W., Yokoo, T. et al. The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nature Plants 4, 1071–1081 (2018). https://doi.org/10.1038/s41477-018-0317-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0317-4
This article is cited by
-
Modulation of the wheat transcriptome by TaZFP13D under well-watered and drought conditions
Plant Molecular Biology (2024)
-
PXL1 and SERKs act as receptor–coreceptor complexes for the CLE19 peptide to regulate pollen development
Nature Communications (2023)
-
A phosphoinositide hub connects CLE peptide signaling and polar auxin efflux regulation
Nature Communications (2023)
-
Determining Germ Cells in Flowering Plants: Common Concepts and Molecular Mechanisms Steering Meiocyte Specification and Mode of Cell Division
Journal of Plant Biology (2023)
-
A Dof-CLE circuit controls phloem organization
Nature Plants (2022)