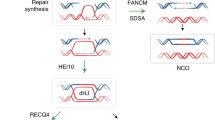
Abstract
Improved plant varieties are important in our attempts to face the challenges of a growing human population and limited planet resources. Plant breeding relies on meiotic crossovers to combine favourable alleles into elite varieties1. However, meiotic crossovers are relatively rare, typically one to three per chromosome2, limiting the efficiency of the breeding process and related activities such as genetic mapping. Several genes that limit meiotic recombination were identified in the model species Arabidopsis thaliana2. Mutation of these genes in Arabidopsis induces a large increase in crossover frequency. However, it remained to be demonstrated whether crossovers could also be increased in crop species hybrids. We explored the effects of mutating the orthologues of FANCM3, RECQ44 or FIGL15 on recombination in three distant crop species, rice (Oryza sativa), pea (Pisum sativum) and tomato (Solanum lycopersicum). We found that the single recq4 mutation increases crossovers about three-fold in these crops, suggesting that manipulating RECQ4 may be a universal tool for increasing recombination in plants. Enhanced recombination could be used with other state-of-the-art technologies such as genomic selection, genome editing or speed breeding6 to enhance the pace and efficiency of plant improvement.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are provided as Supplementary Datasets. Genotyping data and derived recombination frequencies (Figs. 2, 3 and 4) for rice, pea and tomato are given in Supplementary Datasets 2–7, respectively. The protein sequences used for the phylogeny analyses (Supplementary Figs. 1–3) are provided as Supplementary Dataset 1.
References
Wijnker, E. & de Jong, H. Managing meiotic recombination in plant breeding. Trends Plant Sci. 13, 640–646 (2008).
Fernandes, J. B., Seguéla-Arnaud, M., Larchevêque, C., Lloyd, A. H. & Mercier, R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl Acad. Sci. USA 115, 2431–2436 (2018).
Crismani, W. et al. FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012).
Seguéla-Arnaud, M. et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl Acad. Sci. USA 4713–4718 (2015).
Girard, C. et al. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11, e1005369 (2015).
Watson, A. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29 (2018).
Berchowitz, L. E. & Copenhaver, G. P. Genetic interference: don’t stand so close to me. Curr. Genomics 11, 91–102 (2010).
Ziolkowski, P. A. et al. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 31, 306–317 (2017).
Seguéla-Arnaud, M. et al. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 45, 1860–1871 (2017).
Girard, C. et al. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 42, 9087–9095 (2014).
Serra, H. et al. Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl Acad. Sci. USA 115, 2437–2442 (2018).
Hartung, F. & Puchta, H. The RecQ gene family in plants. J. Plant Physiol. 163, 287–296 (2006).
Hartung, F., Suer, S. & Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 104, 18836–18841 (2007).
Ziolkowski, P. A. et al. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife 4, 1–29 (2015).
Blary, A. et al. FANCM limits meiotic crossovers in brassica crops. Front. Plant Sci. 9, 1–13 (2018).
Zhang, P. et al. The rice AAA-ATPase OsFIGNL1 is essential for male meiosis. Front. Plant Sci. 8, 1639 (2017).
Chase, M. W. et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20 (2016).
Lloyd, A. H. et al. Meiotic gene evolution: can you teach a new dog new tricks?. Mol. Biol. Evol. 31, 1724–1727 (2014).
Sallaud, C. et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 39, 450–464 (2004).
Larmande, P. et al. Oryza Tag Line, a phenotypic mutant database for the Génoplante rice insertion line library. Nucleic Acids Res. 36, 1022–1027 (2008).
Zhou, T. et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271, 402–415 (2004).
Alves-Carvalho, S. et al. Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 84, 1–19 (2015).
Fernandes, J. B. et al. FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet. 14, e1007317 (2018).
Zapata, L. et al. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl Acad. Sci. USA 113, E4052–E4060 (2016).
Phillips, D. et al. The effect of temperature on the male and female recombination landscape of barley. New Phytol. 208, 421–429 (2015).
Francis, et al. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl Acad. Sci. USA 104, 3913–3918 (2007).
Lloyd, A. H., Morgan, C., Franklin, C. & Bomblies, K. Plasticity of meiotic recombination rates in response to temperature in . Arabidopsis. Genetics 4, 1409–1420 (2018).
Modliszewski, J. L. et al. Elevated temperature increases meiotic crossover frequency via the interfering (Type I) pathway in Arabidopsis thaliana. PLoS Genet. 14, e1007384 (2018).
Wilfert, L., Gadau, J. & Schmid-Hempel, P. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity 98, 189–197 (2007).
Kwon, Y.-I. et al. DNA replication arrest leads to enhanced homologous recombination and cell death in meristems of rice OsRecQl4 mutants. BMC Plant Biol. 13, 62 (2013).
Nambiar, M. & Smith, G. R. Repression of harmful meiotic recombination in centromeric regions. Semin. Cell Dev. Biol. 54, 188–197 (2016).
Choulet, F. et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721 (2014).
King, J. et al. A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotechnol. J. 15, 217–226 (2017).
Zickler, D. & Kleckner, N. E. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999).
Yin, K., Gao, C. & Qiu, J.-L. Progress and prospects in plant genome editing. Nat. Plants 3, 17107 (2017).
Van Bel, M. et al. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46, D1190–D1196 (2018).
Fernandez-Pozo, N. et al. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041 (2015).
Dereeper, A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 (2008).
Introduction to GATK Best Practices (Broad Institute, 2009); https://software.broadinstitute.org/gatk/best-practices.
Droc, G. et al. OryGenesDB: a database for rice reverse genetics. Nucleic Acids Res. 34, D736–D7340 (2006).
An, S. et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 133, 2040–2047 (2003).
Droc, G., Périn, C., Fromentin, S. & Larmande, P. OryGenesDB 2008 update: database interoperability for functional genomics of rice. Nucleic Acids Res. 37, D992–D995 (2009).
Grelon, M., Vezon, D., Gendrot, G. & Pelletier, G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600 (2001).
Heffelfinger, C., Fragoso, C. A. & Lorieux, M. Constructing linkage maps in the genomics era with MapDisto 2.0. Bioinformatics 33, 2224–2225 (2017).
Tayeh, N. et al. Development of two major resources for pea genomics: the GenoPea 13.2K SNP Array and a high-density, high-resolution consensus genetic map. Plant J. 84, 1257–1273 (2015).
Baldet, P. et al. Investigating the role of vitamin C in tomato through TILLING identification of ascorbate-deficient tomato mutants. Plant Biotechnol. J. 30, 309–314 (2013).
Okabe, Y. et al. Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from micro-tom mutant libraries. Plant Cell Physiol. 52, 1994–2005 (2011).
Garcia, V. et al. Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing. Nat. Protoc. 11, 2401–2418 (2016).
Shirasawa, K., Hirakawa, H., Nunome, T., Tabata, S. & Isobe, S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 14, 51–60 (2016).
Petit, J. et al. Analyses of tomato fruit brightness mutants uncover both cutin-deficient and cutin-abundant mutants and a new hypomorphic allele of GDSL lipase. Plant Physiol. 164, 888–906 (2014).
Smith, S. M. & Maughan, P. J. SNP genotyping using KASPar assays. Methods Mol. Biol. 1245, 243–256 (2015).
Lorieux, M. MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30, 1231–1235 (2012).
Bollier, N. et al. At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30, 83–100 (2018).
Acknowledgements
We thank J. Burstin, M. Causse, B. Courtois and C. Mézard for fruitful discussions; P. Sourdille and F. Benyahya for sharing wheat sequences before publication; C. Le Signor and M.-C. Le Paslier for offering their expertise and advice; and J.-F. Rami for his help with the SpiderMap software. The pea and tomato work was funded by HyperRec grants from INRA Transfert. The Institute Jean-Pierre Bourgin benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). This work was partly funded by the Investissements d’Avenir, France Génomique (10-INBS-0009) project IRIGIN (International Rice Genome Initiative) and the CGIAR research program on rice (RICE).
Author information
Authors and Affiliations
Contributions
D.M., G.A., C.B., A.K., G.D., E.V., C.R.C., M.S., M.D. and J.P.M. produced the data. D.M., G.A., C.B., A.K., E.G. and R.M. analysed the data. D.M., G.A., C.B., C.R., E.G. and R.M. conceived and designed the experiments. D.M. and R.M. wrote the paper with inputs from G.A., C.B., C.R. and E.G.
Corresponding author
Ethics declarations
Competing interests
Patents have been deposited by INRA on the use of RECQ4, FIGL1 and FANCM to manipulate meiotic recombination (EP3149027, EP3016506 and EP2755995).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–9 and Supplementary Tables 1–3.
Dataset 1
Sequences and accession numbers of RECQ4, FIGL1 and FANCM proteins analysed in Supplementary Figures 1–3.
Dataset 2
Genotyping data of rice populations.
Dataset 3
Recombination data in rice.
Dataset 4
Genotyping data of pea populations.
Dataset 5
Recombination data in pea.
Dataset 6
Genotyping data of tomato populations.
Dataset 7
Recombination data in tomato.
Rights and permissions
About this article
Cite this article
Mieulet, D., Aubert, G., Bres, C. et al. Unleashing meiotic crossovers in crops. Nature Plants 4, 1010–1016 (2018). https://doi.org/10.1038/s41477-018-0311-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0311-x
This article is cited by
-
Why do plants need the ZMM crossover pathway? A snapshot of meiotic recombination from the perspective of interhomolog polymorphism
Plant Reproduction (2023)
-
Repair of DNA double-strand breaks in plant meiosis: role of eukaryotic RecA recombinases and their modulators
Plant Reproduction (2023)
-
Technology-driven approaches for meiosis research in tomato and wild relatives
Plant Reproduction (2023)
-
Molecular regulation of tomato male reproductive development
aBIOTECH (2023)
-
Making headway toward enduring changes: perspectives on breeding tree crops through genome editing
Tree Genetics & Genomes (2023)