Abstract
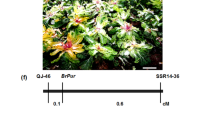
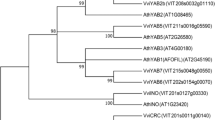
The evolution of fruit colour in plants is intriguing. Citrus fruit has repeatedly gained or lost the ability to synthesize anthocyanins. Chinese box orange, a primitive citrus, can accumulate anthocyanins both in its fruits and its leaves. Wild citrus can accumulate anthocyanins in its leaves. In contrast, most cultivated citrus have lost the ability to accumulate anthocyanins. We characterized a novel MYB regulatory gene, Ruby2, which is adjacent to Ruby1, a known anthocyanin activator of citrus. Different Ruby2 alleles can have opposite effects on the regulation of anthocyanin biosynthesis. AbRuby2Full encodes an anthocyanin activator that mainly functions in the pigmented leaves of Chinese box orange. CgRuby2Short was identified in purple pummelo and encodes an anthocyanin repressor. CgRuby2Short has lost the ability to activate anthocyanin biosynthesis. However, it retains the ability to interact with the same partner, CgbHLH1, as CgRuby1, thus acting as a passive competitor in the regulatory complex. Further investigation in different citrus species indicated that the Ruby2–Ruby1 cluster exhibits subfunctionalization among primitive, wild and cultivated citrus. Our study elucidates the regulatory mechanism and evolutionary history of the Ruby2–Ruby1 cluster in citrus, which are unique and different from that found in Arabidopsis, grape or petunia.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA-seq data have been deposited in NCBI under accession codes SRR7631524 (PP1), SRR7631526 (PP2), SRR7631531 (PP3), SRR7631532 (NP1), SRR7631692 (NP2) and SRR7631800 (NP3).
References
Swingle, W. T. & Reece, P. C. in The Citrus Industry Vol. 1 (eds Reuther, W. et al) 190–430 (Univ. of California Press, Berkeley, 1967).
Butelli, E. et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255 (2012).
Butelli, E. et al. Changes in anthocyanin production during domestication of Citrus. Plant Physiol. 4, 2225–2242 (2017).
Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493 (2001).
Ramsay, N. A. & Glover, B. J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends. Plant. Sci. 10, 63–70 (2005).
Koes, R., Verweij, W. & Quattrocchio, F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends. Plant. Sci. 10, 236–242 (2005).
Xu, W., Dubos, C. & Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends. Plant. Sci. 20, 176–185 (2015).
Albert, N. W. et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980 (2014).
Walker, A. R. et al. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785 (2007).
Espley, R. V. et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21, 168–183 (2009).
Wang, Z. et al. The methylation of the PcMYB10 promoter is associated with green-skinned sport in ‘Max Red Bartlett’ pear. Plant Physiol. 162, 885–896 (2013).
Uematsu, C. et al. Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. J. Exp. Bot. 65, 1081–1094 (2014).
Telias, A. et al. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 11, 93–107 (2011).
Xu, Q. et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45, 59–66 (2012).
Wu, G. A. et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662 (2014).
Wu, G. A. et al. Genomics of the origin and evolution of Citrus. Nature 554, 311–316 (2018).
Hawkins, C., Caruana, J., Schiksnis, E. & Liu, Z. Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep. 6, 29017 (2016).
Wang, X. et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 49, 765–772 (2017).
Lin-Wang, K. et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10, 50–67 (2010).
Chen, J., Hu, Q., Zhang, Y., Lu, C. & Kuang, H. P-MITE: a database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 42, D1176–D1181 (2014).
Zimmermann, I. M., Heim, M. A., Weisshaar, B. & Uhrig, J. F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40, 22–34 (2004).
Hichri, I. et al. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523 (2010).
Gabrielsen, O., Sentenac, A. & Fromageot, P. Specific DNA binding by c-Myb: evidence for a double helix-turn-helix-related motif. Science 253, 1140–1143 (1991).
Jia, L., Clegg, M. T. & Jiang, T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134, 575–585 (2004).
Wright, S. Evolution and Genetics of Populations Vol. 4 (Univ. Chicago Press, Chicago, USA, 1978).
Aharoni, A. et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332 (2001).
Dubos, C. et al. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953 (2008).
Zhu, H. F., Fitzsimmons, K., Khandelwal, A. & Kranz, R. G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2, 790–802 (2009).
Jun, J. H., Liu, C., Xiao, X. & Dixon, R. A. The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell 27, 2860–2879 (2015).
Duarte, J. M. et al. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 23, 469–478 (2006).
Zhang, H. et al. Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants. 2, 16016 (2016).
Gonzalez, A., Zhao, M., Leavitt, J. M. & Lloyd, A. M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 (2008).
Bombarely, A. et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants. 2, 16074 (2016).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome. Biol. 14, R36 (2013).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Hoffmann, T., Kalinowski, G. & Schwab, W. RNAi-induced silencing of gene expression in strawberry fruit (Fragaria x ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J. 48, 818–826 (2006).
Gruntman, E. et al. Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9, 1–14 (2008).
Liu, C. et al. Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 6, 25352 (2016).
Hellens, R. P. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005).
Cao, H. et al. Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J. Exp. Bot. 63, 4403–4417 (2012).
Zhu, F. et al. An R2R3-MYB transcription factor represses the transformation of alpha- and beta-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 216, 178–192 (2017).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Acknowledgements
We thank C.Y. Kang (Huazhong Agricultural University) for kindly providing the strawberry fruits for transient expression assay, L.Z. Xiong and W. Zong (Huazhong Agricultural University) for providing plasmids pM999-35 and 35S:Ghd7–CFP and A. Allan (The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand) for providing the vectors for Dual Luciferase Assay. This project was supported by the National Natural Science Foundation of China (31872052, 31572105 and 31330066 to Q.X. and X.X.D.), the fundamental research funds for the central universities (2662015PY109 and 2662018PY008 to Q.X.) and Huazhong Agricultural University Scientific & Technological Self-innovation Foundation.
Author information
Authors and Affiliations
Contributions
Q.X. conceived and coordinated this project. D.H designed the experiments. D.H., Z.Z.T., Y.T.X., X.L.J, Y.Y. and J.X.H, performed the experiments. X.X.D. and S.A.P found and collected the purple pummelo. D.H. and X.W. analysed the bioinformatic data. L.L. and E.B. were involved in the research design and the improvement of the manuscript. D.H. and Q.X. wrote the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–16 and Supplementary Tables 2–6.
Supplementary Table 1
Gene expression profiling in the fruit peel was compared between the purple pummelo (PP) and normal pummelo (NP).
Supplementary Table 7
List of oligonucleotide sequences used in this study.
Supplementary Data Set 1
Nucleotide sequence of Ruby2 alleles in different accessions of the genus Citrus.
Supplementary Data Set 2
Re-sequencing of Ruby1 promoter in different accessions of the genus Citrus.
Supplementary Data Set 3
SNPs from the 11 accessions in the regions of single-copy genes that are conserved among Citrinae genomes.
Rights and permissions
About this article
Cite this article
Huang, D., Wang, X., Tang, Z. et al. Subfunctionalization of the Ruby2–Ruby1 gene cluster during the domestication of citrus. Nature Plants 4, 930–941 (2018). https://doi.org/10.1038/s41477-018-0287-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0287-6
This article is cited by
-
Biofortified Rice Provides Rich Sakuranetin in Endosperm
Rice (2024)
-
Genome-wide characterization of the bHLH gene family in Gynostemma pentaphyllum reveals its potential role in the regulation of gypenoside biosynthesis
BMC Plant Biology (2024)
-
Haplotype-resolved genome assembly of the diploid Rosa chinensis provides insight into the mechanisms underlying key ornamental traits
Molecular Horticulture (2024)
-
Transcriptomics and metabolomics reveal the underlying mechanism of drought treatment on anthocyanin accumulation in postharvest blood orange fruit
BMC Plant Biology (2024)
-
Molecular characterization of a flavanone 3-hydroxylase gene from citrus fruit reveals its crucial roles in anthocyanin accumulation
BMC Plant Biology (2023)