Abstract
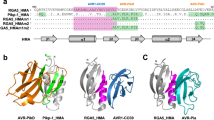
Accelerated adaptive evolution is a hallmark of plant–pathogen interactions. Plant intracellular immune receptors (NLRs) often occur as allelic series with differential pathogen specificities. The determinants of this specificity remain largely unknown. Here, we unravelled the biophysical and structural basis of expanded specificity in the allelic rice NLR Pik, which responds to the effector AVR-Pik from the rice blast pathogen Magnaporthe oryzae. Rice plants expressing the Pikm allele resist infection by blast strains expressing any of three AVR-Pik effector variants, whereas those expressing Pikp only respond to one. Unlike Pikp, the integrated heavy metal-associated (HMA) domain of Pikm binds with high affinity to each of the three recognized effector variants, and variation at binding interfaces between effectors and Pikp-HMA or Pikm-HMA domains encodes specificity. By understanding how co-evolution has shaped the response profile of an allelic NLR, we highlight how natural selection drove the emergence of new receptor specificities. This work has implications for the engineering of NLRs with improved utility in agriculture.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
20 August 2018
In the version of this Article originally published, in Fig. 1b the single-letter code for the amino acid polymorphism at position 46 in the schematic of the AVR-PikE variant was incorrectly given as ‘H’. The correct amino acid is ‘N’. This has now been amended in all versions of the Article.
References
Jones, J. D., Vance, R. E. & Dangl, J. L. Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 (2016).
Ronald, P. C. & Beutler, B. Plant and animal sensors of conserved microbial signatures. Science 330, 1061–1064 (2010).
Dodds, P. N. & Rathjen, J. P. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010).
Win, J. et al. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 235–247 (2012).
Bialas, A. et al. Lessons in effector and NLR biology of plant–microbe systems. Mol. Plant Microbe Interact. 31, 34–45 (2017).
Ellis, J. G., Lawrence, G. J., Luck, J. E. & Dodds, P. N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506 (1999).
Allen, R. L. et al. Host–parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960 (2004).
Bhullar, N. K., Zhang, Z., Wicker, T. & Keller, B. Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biol. 10, 88 (2010).
Seeholzer, S. et al. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant Microbe Interact. 23, 497–509 (2010).
Srichumpa, P., Brunner, S., Keller, B. & Yahiaoui, N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 139, 885–895 (2005).
Lu, X. et al. Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl Acad. Sci. USA 113, E6486–E6495 (2016).
Dodds, P. N. et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl Acad. Sci. USA 103, 8888–8893 (2006).
Krasileva, K. V., Dahlbeck, D. & Staskawicz, B. J. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22, 2444–2458 (2010).
Steinbrenner, A. D., Goritschnig, S. & Staskawicz, B. J. Recognition and activation domains contribute to allele-specific responses of an Arabidopsis NLR receptor to an oomycete effector protein. PLoS Pathog. 11, e1004665 (2015).
Wang, C. I. et al. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 19, 2898–2912 (2007).
Huang, J., Si, W., Deng, Q., Li, P. & Yang, S. Rapid evolution of avirulence genes in rice blast fungus Magnaporthe oryzae. BMC Genet. 15, 45 (2014).
Raffaele, S. et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330, 1540–1543 (2010).
Yoshida, K. et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591 (2009).
Dangl, J. L., Horvath, D. M. & Staskawicz, B. J. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 (2013).
Rodriguez-Moreno, L., Song, Y. & Thomma, B. P. Transfer and engineering of immune receptors to improve recognition capacities in crops. Curr. Opin. Plant Biol. 38, 42–49 (2017).
Eitas, T. K. & Dangl, J. L. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13, 472–477 (2010).
Wu, C. H., Belhaj, K., Bozkurt, T. O., Birk, M. S. & Kamoun, S. Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. New Phytol. 209, 1344–1352 (2016).
Narusaka, M. et al. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226 (2009).
Sinapidou, E. et al. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38, 898–909 (2004).
Ashikawa, I. et al. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180, 2267–2276 (2008).
Lee, S. K. et al. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181, 1627–1638 (2009).
Wu, C. H. et al. NLR network mediates immunity to diverse plant pathogens. Proc. Natl Acad. Sci. USA 114, 8113–8118 (2017).
Cesari, S., Bernoux, M., Moncuquet, P., Kroj, T. & Dodds, P. N. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 5, 606 (2014).
Kroj, T., Chanclud, E., Michel-Romiti, C., Grand, X. & Morel, J. B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626 (2016).
Sarris, P. F., Cevik, V., Dagdas, G., Jones, J. D. & Krasileva, K. V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14, 8 (2016).
Wu, C. H., Krasileva, K. V., Banfield, M. J., Terauchi, R. & Kamoun, S.The “sensor domains” of plant NLR proteins: more than decoys?. Front. Plant Sci. 6, 134 (2015).
Okuyama, Y. et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66, 467–479 (2011).
Maqbool, A. et al. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4, e08709 (2015).
Kanzaki, H. et al. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907 (2012).
Costanzo, S. & Jia, Y. L. Sequence variation at the rice blast resistance gene Pi-km locus: implications for the development of allele specific markers. Plant Sci. 178, 523–530 (2010).
Krissinel, E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43, W314–W319 (2015).
de Guillen, K. et al. Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 11, e1005228 (2015).
Bent, A. F. et al. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860 (1994).
Mindrinos, M., Katagiri, F., Yu, G. L. & Ausubel, F. M. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099 (1994).
Whitham, S. et al. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115 (1994).
Le Roux, C. et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161, 1074–1088 (2015).
Sarris, P. F. et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100 (2015).
Zhang, Z. M. et al. Mechanism of host substrate acetylation by a YopJ family effector. Nat. Plants 3, 17115 (2017).
Ortiz, D. et al. Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 29, 156–168 (2017).
Lobstein, J. et al. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 11, 56 (2012).
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Wickham, H. ggplot. Elegant Graphics for Data Analysis (Springer, New York, NY, 2009).
Winter, G. xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
This work was supported by the BBSRC (grants BB/J00453, BB/P012574 and BB/M02198X), the ERC (proposal 743165), the John Innes Foundation, the Gatsby Charitable Foundation and JSPS KAKENHI 15H05779. We thank the Diamond Light Source (beamlines I03 and I04 under proposals MX9475 and MX13467) for access to X-ray data collection facilities. We also thank D. Lawson and C. Stevenson (JIC X-ray Crystallography/Biophysical Analysis Platform) for help with protein structure determination and SPR.
Author information
Authors and Affiliations
Contributions
J.C.D.l.C. and M.F. performed all of the experiments. J.C.D.l.C., M.F. and M.J.B. designed the experiments and analysed the data. A.M. and H.S. assisted with construct design and the initial protein production. R.T. and S.K. analysed the data. J.C.D.l.C., M.F. and M.J.B. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Figures 1–5, Supplementary Tables 1–4 and Supplementary References
Rights and permissions
About this article
Cite this article
De la Concepcion, J.C., Franceschetti, M., Maqbool, A. et al. Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nature Plants 4, 576–585 (2018). https://doi.org/10.1038/s41477-018-0194-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0194-x
This article is cited by
-
Early molecular events in the interaction between Magnaporthe oryzae and rice
Phytopathology Research (2024)
-
An pair of an atypical NLR encoding genes confer Asian soybean rust resistance in soybean
Nature Communications (2024)
-
The synthetic NLR RGA5HMA5 requires multiple interfaces within and outside the integrated domain for effector recognition
Nature Communications (2024)
-
Prediction of effector protein structures from fungal phytopathogens enables evolutionary analyses
Nature Microbiology (2023)
-
The Venturia inaequalis effector repertoire is dominated by expanded families with predicted structural similarity, but unrelated sequence, to avirulence proteins from other plant-pathogenic fungi
BMC Biology (2022)