Abstract
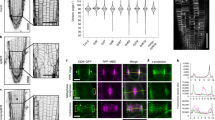
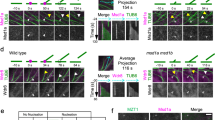
The evolutionarily conserved WD40 protein budding uninhibited by benzimidazole 3 (BUB3) is known for its function in spindle assembly checkpoint control. In the model plant Arabidopsis thaliana, nearly identical BUB3;1 and BUB3;2 proteins decorated the phragmoplast midline through interaction with the microtubule-associated protein MAP65-3 during cytokinesis. BUB3;1 and BUB3;2 interacted with the carboxy-terminal segment of MAP65-3 (but not MAP65-1), which harbours its microtubule-binding domain for its post-mitotic localization. Reciprocally, BUB3;1 and BUB3;2 also regulated MAP65-3 localization in the phragmoplast by enhancing its microtubule association. In the bub3;1 bub3;2 double mutant, MAP65-3 localization was often dissipated away from the phragmoplast midline and abolished upon treatment of low doses of the cytokinesis inhibitory drug caffeine that were tolerated by the control plant. The phragmoplast microtubule array exhibited uncoordinated expansion pattern in the double mutant cells as the phragmoplast edge reached the parental plasma membrane at different times in different areas. Upon caffeine treatment, phragmoplast expansion was halted as if the microtubule array was frozen. As a result, cytokinesis was abolished due to failed cell plate assembly. Our findings have uncovered a novel function of the plant BUB3 in MAP65-3-dependent microtubule reorganization during cytokinesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
07 August 2018
In the version of this Article originally published, the affiliation for author Yuh-Ru Julie Lee was incorrect; the correct affiliation is ‘2Department of Plant Biology, College of Biological Sciences, University of California, Davis, CA, USA’. This has now been amended in all versions of the Article.
References
Hoyt, M. A., Totis, L. & Roberts, B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66, 507–517 (1991).
Musacchio, A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25, R1002–R1018 (2015).
Li, R. & Murray, A. W. Feedback control of mitosis in budding yeast. Cell 66, 519–531 (1991).
Babu, J. R. et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160, 341–353 (2003).
Komaki, S. & Schnittger, A. The spindle assembly checkpoint in Arabidopsis is rapidly shut off during severe stress. Dev. Cell 43, 172–185.e5 (2017).
Lermontova, I., Fuchs, J. & Schubert, I. The Arabidopsis checkpoint protein Bub3.1 is essential for gametophyte development. Front Biosci. 13, 5202–5211 (2008).
Van Leene, J. et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6, 397 (2010).
Smertenko, A. P. et al. The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 20, 3346–3358 (2008).
Müller, S. et al. The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr. Biol. 14, 412–417 (2004).
Ho, C. M. et al. Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell 23, 2909–2923 (2011).
Ho, C. M., Lee, Y. R., Kiyama, L. D., Dinesh-Kumar, S. P. & Liu, B. Arabidopsis microtubule-associated protein MAP65-3 cross-links antiparallel microtubules toward their plus ends in the phragmoplast via its distinct C-terminal microtubule binding domain. Plant Cell 24, 2071–2085 (2012).
Smertenko, A. P. et al. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 16, 2035–2047 (2004).
Lee, Y. J. et al. The mitotic function of augmin is dependent on its microtubule-associated protein subunit EDE1 in Arabidopsis thaliana. Curr. Biol. 27, 3891–3897.e4 (2017).
Hepler, P. K. & Bonsignore, C. L. Caffeine inhibition of cytokinesis—ultrastructure of cell plate formation degradation. Protoplasma 157, 182–192 (1990).
Kang, B. H., Busse, J. S. & Bednarek, S. Y. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913 (2003).
Smertenko, A. et al. Plant cytokinesis: terminology for structures and processes. Trends Cell Biol. 27, 885–894 (2017).
Eleftheriou, E. P., Baskin, T. I. & Hepler, P. K. Aberrant cell plate formation in the Arabidopsis thaliana microtubule organization 1 mutant. Plant Cell Physiol. 46, 671–675 (2005).
Steiner, A. et al. The membrane-associated Sec1/Munc18 KEULE is required for phragmoplast microtubule reorganization during cytokinesis in Arabidopsis. Mol. Plant 9, 528–540 (2016).
Steiner, A. et al. Cell cycle-regulated PLEIADE/AtMAP65-3 links membrane and microtubule dynamics during plant cytokinesis. Plant J. 88, 531–541 (2016).
Waizenegger, I. et al. The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr. Biol. 10, 1371–1374 (2000).
Lee, Y. R. & Liu, B. The rise and fall of the phragmoplast microtubule array. Curr. Opin. Plant Biol. 16, 757–763 (2013).
London, N. & Biggins, S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15, 736–747 (2014).
Ding, D., Muthuswamy, S. & Meier, I. Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol. Biol. 79, 203–216 (2012).
Komaki, S. & Schnittger, A. The spindle checkpoint in plants—a green variation over a conserved theme? Curr. Opin. Plant Biol. 34, 84–91 (2016).
Van Damme, D. et al. Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23, 4013–4024 (2011).
Efimov, V. P. & Morris, N. R. A screen for dynein synthetic lethals in Aspergillus nidulans identifies spindle assembly checkpoint genes and other genes involved in mitosis. Genetics 149, 101–116 (1998).
Bao, Z., Zhang, N. & Hua, J. Endopolyploidization and flowering time are antagonistically regulated by checkpoint component MAD1 and immunity modulator MOS1. Nat. Commun. 5, 5628 (2014).
Kevei, Z. et al. Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS ONE 6, e20618 (2011).
Tange, Y. & Niwa, O. Schizosaccharomyces pombe Bub3 is dispensable for mitotic arrest following perturbed spindle formation. Genetics 179, 785–792 (2008).
Caillaud, M. C. et al. Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS ONE 4, e6757 (2009).
Yu, H. G., Muszynski, M. G. & Kelly Dawe, R. The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 145, 425–435 (1999).
Hu, C. K., Ozlu, N., Coughlin, M., Steen, J. J. & Mitchison, T. J. Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol. Biol. Cell 23, 2702–2711 (2012).
Smertenko, A. P. et al. Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J. Cell Sci. 119, 3227–3237 (2006).
Sasabe, M. & Machida, Y. Regulation of organization and function of microtubules by the mitogen-activated protein kinase cascade during plant cytokinesis. Cytoskeleton (Hoboken) 69, 913–918 (2012).
Nishihama, R. et al. Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109, 87–99 (2002).
Kosetsu, K. et al. The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22, 3778–3790 (2010).
Murata, T. et al. Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat. Commun. 4, 1967 (2013).
Li, H. et al. Arabidopsis MAP65-4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol. 215, 187–201 (2017).
Ravikumar, R., Steiner, A. & Assaad, F. F. Multisubunit tethering complexes in higher plants. Curr. Opin. Plant Biol. 40, 97–105 (2017).
Sasabe, M., Kosetsu, K., Hidaka, M., Murase, A. & Machida, Y. Arabidopsis thaliana MAP65-1 and MAP65-2 function redundantly with MAP65-3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal. Behav. 6, 743–747 (2011).
Kong, Z., Hotta, T., Lee, Y. R., Horio, T. & Liu, B. The γ-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell 22, 191–204 (2010).
Nakamura, S. et al. Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 74, 1315–1319 (2010).
Nakagawa, T. et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 (2007).
Lee, Y. R. J. & Liu, B. Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr. Biol. 10, 797–800 (2000).
Edelstein, A., Amodaj, N., Hoover, K. & Vale, R. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. 92, 14.20.1–14.20.17 (2010).
Edelstein, A. D. et al. Advanced methods of microscope control using µManager software. J. Biol. Methods 1, e10 (2014).
Acknowledgements
We thank members of the Liu laboratory for critical comments on the work. Special thanks go to S. P. Dinesh-Kumar for the BiFC vectors and T. Nakagawa at Shimane University in Japan for the pGWB plasmids, and S. Bednarek for the DRP1A antibody. This work was supported by the US National Science Foundation under the grant MCB-1412509 to B.L. and Y.-R.J.L. H.Z. was supported by a fellowship from the China Scholarship Council (no. 201406305055), the Fundamental Research Funds for the Central Universities (no. 2452018155) and the 111 Project from the Ministry of Education of China (no. B07049). Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agency.
Author information
Authors and Affiliations
Contributions
Y.-R.J.L. and B.L. conceived the project and designed the experiments. H.Z., X.D., B.S., S.L.V. and Y.-R.J.L. performed the experiments. H.Z., X.D., Z.K., H.L., Y.-R.J.L. and B.L. analysed the data and interpreted the results. Y.-R.J.L. and B.L. wrote the manuscript with inputs from other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, and Supplementary Figures 1 and 2
Supplementary Video 1
BUB3;1-GFP localization during mitotic cell division in A. thaliana. This experiment was repeated independently five times with similar results.
Supplementary Video 2
MT reorganization during mitotic cell division in a control cell (left) and a bub3;1 bub3;2 double mutant cell (right) in A. thaliana under undisturbed conditions. This experiment was repeated independently three times with similar results.
Supplementary Video 3
MT reorganization during mitotic cell division in a control cell in A. thaliana after being exposed to 1 mM caffeine this experiment was repeated independently three times with similar results.
Supplementary Video 4
MT reorganization during mitotic cell division in a bub3;1 bub3;2 double mutant cell in A. thaliana after being exposed to 1 mM caffeine. This experiment was repeated independently three times with similar results.
Rights and permissions
About this article
Cite this article
Zhang, H., Deng, X., Sun, B. et al. Role of the BUB3 protein in phragmoplast microtubule reorganization during cytokinesis. Nature Plants 4, 485–494 (2018). https://doi.org/10.1038/s41477-018-0192-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0192-z
This article is cited by
-
Comprehensive analyses of microtubule-associated protein MAP65 family genes in Cucurbitaceae and CsaMAP65s expression profiles in cucumber
Journal of Applied Genetics (2023)