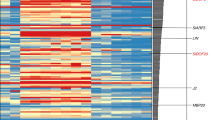
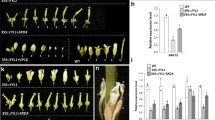
Abstract
Flowers have a species-specific functional life span that determines the time window in which pollination, fertilization and seed set can occur. The stigma tissue plays a key role in flower receptivity by intercepting pollen and initiating pollen tube growth toward the ovary. In this article, we show that a developmentally controlled cell death programme terminates the functional life span of stigma cells in Arabidopsis. We identified the leaf senescence regulator ORESARA1 (also known as ANAC092) and the previously uncharacterized KIRA1 (also known as ANAC074) as partially redundant transcription factors that modulate stigma longevity by controlling the expression of programmed cell death–associated genes. KIRA1 expression is sufficient to induce cell death and terminate floral receptivity, whereas lack of both KIRA1 and ORESARA1 substantially increases stigma life span. Surprisingly, the extension of stigma longevity is accompanied by only a moderate extension of flower receptivity, suggesting that additional processes participate in the control of the flower’s receptive life span.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rogers, H. J. From models to ornamentals: how is flower senescence regulated?. Plant Mol. Biol. 82, 563–574 (2013).
Jones, M. L. Ethylene signaling is required for pollination-accelerated corolla senescence in petunias. Plant Sci. 175, 190–196 (2008).
Williams, R. R. The effect of summer nitrogen applications on the quality of apple blossom. J. Horticult. Sci. 40, 31–41 (1965).
Shahri, W. & Tahir, I. Flower senescence: some molecular aspects. Planta 239, 277–297 (2014).
Shibuya, K., Yamada, T., & Ichimura, K. Morphological changes in senescing petal cells and the regulatory mechanism of petal senescence. J. Exp. Bot. 67, 5909–5918 (2016).
Broderick, S. R. et al. RNA-sequencing reveals early, dynamic transcriptome changes in the corollas of pollinated petunias. BMC Plant Biol. 14, 307 (2014).
Wagstaff, C., Yang, T. J., Stead, A. D., Buchanan-Wollaston, V., & Roberts, J. A. A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J. 57, 690–705 (2009).
Shibuya, K., Shimizu, K., Niki, T. & Ichimura, K. Identification of a NAC transcription factor, EPHEMERAL1, that controls petal senescence in Japanese morning glory. Plant J. 79, 1044–1051 (2014).
Kim, H. J. et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036 (2014).
Kim, J. H. et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057 (2009).
Dickman, M., Williams, B., Li, Y., de Figueiredo, P., & Wolpert, T. Reassessing apoptosis in plants. Nat. Plants 3, 773–779 (2017).
Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 197, 696–711 (2013).
Daneva, A., Gao, Z., Van Durme, M. & Nowack, M. K. Functions and regulation of programmed cell death in plant development. Annu Rev. Cell Dev. Biol. 32, 441–468 (2016).
Huysmans, M., Lema, A. S., Coll, N. S. & Nowack, M. K. Dying two deaths – programmed cell death regulation in development and disease. Curr. Opin. Plant Biol. 35, 37–44 (2017).
Olvera-Carrillo, Y. et al. A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiol. 169, 2684–2699 (2015).
Heslop-Harrison, Y. & Shivanna, K. R. The receptive surface of the angiosperm stigma. Ann. Bot. 41, 1233–1258 (1977).
Dresselhaus, T. & Franklin-Tong, N. Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018–1036 (2013).
Christensen, C. A., King, E. J., Jordan, J. R. & Drews, G. N. Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10, 49–64 (1997).
Weijers, D., Van Hamburg, J. P., Van Rijn, E., Hooykaas, P. J. & Offringa, R. Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 133, 1882–1892 (2003).
Hackett, R. M., Cadwallader, G. & Franklin, F. C. Functional analysis of a Brassica oleracea SLR1 gene promoter. Plant Physiol. 112, 1601–1607 (1996).
Thorsness, M. K., Kandasamy, M. K., Nasrallah, M. E. & Nasrallah, J. B. Genetic ablation of floral cells in Arabidopsis. Plant Cell 5, 253–261 (1993).
Fendrych, M. et al. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 24, 931–940 (2014).
Jones, K., Kim, D. W., Park, J. S. & Khang, C. H. Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biol. 16, 69 (2016).
Kim, H. J., Nam, H. G., & Lim, P. O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56 (2016).
Balazadeh, S. et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264 (2010).
He, X. J. et al. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916 (2005).
Hiratsu, K., Matsui, K., Koyama, T., & Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 (2003).
Mitsuda, N. et al. CRES-T, an effective gene silencing system utilizing chimeric repressors. Methods Mol. Biol. 754, 87–105 (2011).
Zhou, L. Z. et al. Expression analysis of KDEL-CysEPs programmed cell death markers during reproduction in Arabidopsis. Plant Reprod. 29, 265–272 (2016).
Siligato, R. et al. MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 170, 627–641 (2016).
Matallana-Ramirez, L. P. et al. NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 6, 1432–1452 (2013).
Vanden Bossche, R., Demedts, B., Vanderhaeghen, R. & Goossens, A. Transient expression assays in tobacco protoplasts. Methods Mol. Biol. 1011, 227–239 (2013).
Johnson, M. A. et al. Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168, 971–982 (2004).
Carbonell-Bejerano, P., Urbez, C., Carbonell, J., Granell, A. & Perez-Amador, M. A. A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol. 154, 163–172 (2010).
Sanzol, J., & Herrero, M. The “effective pollination period” in fruit trees. Sci. Hortic. 90, 1–17 (2001).
Ohashi-Ito, K., Oda, Y. & Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473 (2010).
van Doorn, W. G. Classes of programmed cell death in plants, compared to those in animals. J. Exp. Bot. 62, 4749–4761 (2011).
van Doorn, W. G. et al. Morphological classification of plant cell deaths. Cell Death Differ. 18, 1241–1246 (2011).
Obara, K., Kuriyama, H. & Fukuda, H. Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol. 125, 615–626 (2001).
Crawford, B. C. & Yanofsky, M. F. HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 138, 2999–3009 (2011).
Ferradás, Y., López, M., Rey, M. & González, M. V. Programmed cell death in kiwifruit stigmatic arms and its relationship to the effective pollination period and the progamic phase. Ann. Bot. 114, 35–45 (2014).
Bac-Molenaar, J. A. et al. Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 27, 1857–1874 (2015).
Franco-Zorrilla, J. M. et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl Acad. Sci. USA 111, 2367–2372 (2014).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Thomas-Chollier, M. et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 36, W119–W127 (2008).
Ritter, A. et al. The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat. Commun. 8, 15235 (2017).
Wong, J. L., Leydon, A. R. & Johnson, M. A. HAP2(GCS1)-dependent gamete fusion requires a positively charged carboxy-terminal domain. PLoS Genet 6, e1000882 (2010).
Leroux, C. et al. PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination. Plant Physiol. 167, 367–380 (2015).
Acknowledgements
We thank the members of the PCD laboratory for discussions and critical feedback on the manuscript, V. Storme for assistance with statistical analysis, and A. Bleys for help with preparing the manuscript. We gratefully acknowledge funding from the Chinese Scholarship Council (CSC; project number 201206910025 to Z.G.), the Fonds Wetenschappelijk Onderzoek (FWO; project number G005112N to A.D.; fellowship number 12I7417N to Z.L.), the Belgian Federal Science Policy Office (BELSPO; to Y.S.), the Agency for Innovation by Science and Technology of Belgium (IWT; fellowship number 121110 to M.V.D.), the Hercules foundation (grant AUGE-09-029 to K.D.), and the ERC StG PROCELLDEATH (project number 639234 to M.K.N.).
Author information
Authors and Affiliations
Contributions
Z.G. and A.D., designed and performed most of the experiments. Y.S. initiated the project and performed experiments. M.V.D., M.H., Z.L., F.D.W., S.V. and M.K. performed experiments, analysed the data and interpreted the results. J.V.d.V. and K.V. performed TF binding site predictions. D.V.d.W. and K.D. contributed to the cryo-scanning electron microscopy experiments, and B.N.L. contributed to the setup of stigma live-cell imaging. M.K.N. supervised the project and designed the experiments. Z.G., A.D. and M.K.N. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–7, Supplementary References, Supplementary Table 2 and Supplementary Methods
Supplementary Table 1
Cluster analysis of differentially expressed genes identified by RNA sequencing of developmental stigma series.
Supplementary Table 3
A table of all primers used in this study.
Supplementary Video 1
Macroscopic phenotyping of an unpollinated stigma life span with an SLR camera.
Supplementary Video 2
Macroscopic phenotyping of an unpollinated stigma life span with the webcam system.
Supplementary Video 3
Microscopic imaging of stigmatic papilla cell death.
Supplementary Video 4
Microscopic imaging of stigmatic papilla cells dying in clusters.
Supplementary Video 5
Microscopic imaging of cell death events taking place in dying papilla cell.
Supplementary Video 6
ORE1 and KIR1 mutants show extended papilla life spans.
Supplementary Video 7
Induced overexpression of KIR1 and ORE1 leads to growth arrest and death of entire seedlings.
Rights and permissions
About this article
Cite this article
Gao, Z., Daneva, A., Salanenka, Y. et al. KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis. Nature Plants 4, 365–375 (2018). https://doi.org/10.1038/s41477-018-0160-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0160-7
This article is cited by
-
Examining preharvest genetic and morphological factors contributing to lettuce (Lactuca sativa L.) shelf-life
Scientific Reports (2024)
-
CAFRI-Arabidopsis: An Intuitive Web-Based Functional Redundancy Inspector in Arabidopsis
Journal of Plant Biology (2024)
-
Exo84c interacts with VAP27 to regulate exocytotic compartment degradation and stigma senescence
Nature Communications (2023)
-
The Arabidopsis SNARE complex genes regulate the early stages of pollen–stigma interactions
Plant Reproduction (2023)
-
Comparative transcriptomics identifies candidate genes involved in the evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae)
BMC Plant Biology (2022)