Abstract
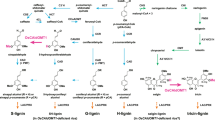
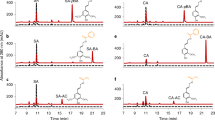
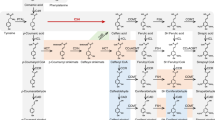
Lignin is a complex and irregular biopolymer of crosslinked phenylpropanoid units in plant secondary cell walls. Its biosynthesis requires three endoplasmic reticulum (ER)-resident cytochrome P450 monooxygenases, C4H, C3ʹH and F5H, to establish the structural characteristics of its monomeric precursors. These P450 enzymes were reported to associate with each other or potentially with other soluble monolignol biosynthetic enzymes to form an enzyme complex or a metabolon. However, the molecular basis governing such enzyme or pathway organization remains elusive. Here, we show that Arabidopsis membrane steroid-binding proteins (MSBPs) serve as a scaffold to physically organize monolignol P450 monooxygenases, thereby regulating the lignin biosynthetic process. We find that although C4H, C3ʹH and F5H are in spatial proximity to each other on the ER membrane in vivo, they do not appear to directly interact with each other. Instead, two MSBP proteins physically interact with all three P450 enzymes and, moreover, MSBPs themselves associate as homomers and heteromers on the ER membrane, thereby organizing P450 clusters. Downregulation of MSBP genes does not affect the transcription levels of monolignol biosynthetic P450 genes but substantially impairs the stability and activity of the MSBP-interacting P450 enzymes and, consequently, lignin deposition, and the accumulation of soluble phenolics in the monolignol branch but not in the flavonoid pathway. Our study suggests that MSBP proteins are essential structural components in the ER membrane that physically organize and stabilize the monolignol biosynthetic P450 enzyme complex, thereby specifically controlling phenylpropanoid–monolignol branch biosynthesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Boerjan, W., Ralph, J. & Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 (2003).
Weng, J. K., Li, X., Bonawitz, N. D. & Chapple, C. Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19, 166–172 (2008).
Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 3, 2–20 (2010).
Jorgensen, K. et al. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8, 280–291 (2005).
Srere, P. A. The metabolon. Trends Biochem. Sci. 10, 109–110 (1985).
Achnine, L., Blancaflor, E. B., Rasmussen, S. & Dixon, R. A. Colocalization of l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16, 3098–3109 (2004).
Bassard, J. E. et al. Protein–protein and protein–membrane associations in the lignin pathway. Plant Cell 24, 4465–4482 (2012).
Rasmussen, S. & Dixon, R. A. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11, 1537–1551 (1999).
Winkel-Shirley, B. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107, 142–149 (1999).
Chen, H. C. et al. Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl Acad. Sci. USA 108, 21253–21258 (2011).
Mifsud, W. & Bateman, A. Membrane-bound progesterone receptors contain a cytochrome b5-like ligand-binding domain. Genome Biol. 3, RESEARCH0068 (2002).
Rohe, H. J., Ahmed, I. S., Twist, K. E. & Craven, R. J. PGRMC1 (progesterone receptor membrane component 1): a targetable protein with multiple functions in steroid signaling, P450 activation and drug binding. Pharmacol. Ther. 121, 14–19 (2009).
Piel, R. B. 3rd et al. A novel role for progesterone receptor membrane component 1 (PGRMC1): a partner and regulator of ferrochelatase. Biochemistry 55, 5204–5217 (2016).
Hand, R. A., Jia, N., Bard, M. & Craven, R. J. Saccharomyces cerevisiae Dap1p, a novel DNA damage response protein related to the mammalian membrane-associated progesterone receptor. Eukaryot. Cell 2, 306–317 (2003).
Mallory, J. C. et al. Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 1669–1679 (2005).
Ghosh, K. et al. Spectroscopic and biochemical characterization of heme binding to yeast Dap1p and mouse PGRMC1p. Biochemistry 44, 16729–16736 (2005).
Kaluka, D., Batabyal, D., Chiang, B. Y., Poulos, T. L. & Yeh, S. R. Spectroscopic and mutagenesis studies of human PGRMC1. Biochemistry 54, 1638–1647 (2015).
Hughes, A. L. et al. Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 5, 143–149 (2007).
Min, L. et al. Molecular identification of adrenal inner zone antigen as a heme-binding protein. FEBS J. 272, 5832–5843 (2005).
Min, L. et al. Characterization of the adrenal-specific antigen IZA (inner zone antigen) and its role in the steroidogenesis. Mol. Cell Endocrinol. 215, 143–148 (2004).
Kabe, Y. et al. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 7, 11030 (2016).
Yang, X. H., Xu, Z. H. & Xue, H. W. Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation. Plant Cell 17, 116–131 (2005).
Song, L., Shi, Q. M., Yang, X. H., Xu, Z. H. & Xue, H. W. Membrane steroid-binding protein 1 (MSBP1) negatively regulates brassinosteroid signaling by enhancing the endocytosis of BAK1. Cell Res. 19, 864–876 (2009).
Yang, X., Song, L. & Xue, H. W. Membrane steroid binding protein 1 (MSBP1) stimulates tropism by regulating vesicle trafficking and auxin redistribution. Mol. Plant 1, 1077–1087 (2008).
Johnsson, N. & Varshavsky, A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA 91, 10340–10344 (1994).
Stagljar, I., Korostensky, C., Johnsson, N. & te Heesen, S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl Acad. Sci. USA 95, 5187–5192 (1998).
Grefen, C., Lalonde, S. & Obrdlik, P. Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr. Protoc. Neurosci. 41, 1–41 (2007).
Obrdlik, P. et al. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl Acad. Sci. USA 101, 12242–12247 (2004).
Dastmalchi, M., Bernards, M. A. & Dhaubhadel, S. Twin anchors of the soybean isoflavonoid metabolon: evidence for tethering of the complex to the endoplasmic reticulum by IFS and C4H. Plant J. 85, 689–706 (2016).
Ozalp, C., Szczesna-Skorupa, E. & Kemper, B. Bimolecular fluorescence complementation analysis of cytochrome p450 2c2, 2e1, and NADPH-cytochrome p450 reductase molecular interactions in living cells. Drug Metab. Dispos. 33, 1382–1390 (2005).
Sundin, L. et al. Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol. 166, 1956–1971 (2014).
Qi, Y. & Katagiri, F. Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 57, 932–944 (2009).
Matsushima, R. et al. An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130, 1807–1814 (2002).
Larsson, C., Sommarin, M. & Widell, S. Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Methods Enzymol. 228, 451–496 (1994).
Zhang, X., Gou, M. & Liu, C. J. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25, 4994–5010 (2013).
Mitsuda, N. & Ohme-Takagi, M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 56, 768–778 (2008).
Ryu, C. S., Klein, K. & Zanger, U. M. Membrane associated progesterone receptors: promiscuous proteins with pleiotropic functions - focus on interactions with cytochromes P450. Front. Pharmacol. 8, 159 (2017).
Backes, W. L. & Kelley, R. W. Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol. Ther. 98, 221–233 (2003).
Kelley, R. W., Cheng, D. & Backes, W. L. Heteromeric complex formation between CYP2E1 and CYP1A2: evidence for the involvement of electrostatic interactions. Biochemistry 45, 15807–15816 (2006).
Scott, E. E. et al. The role of protein–protein and protein–membrane interactions on P450 function. Drug Metab. Dispos. 44, 576–590 (2016).
Kerppola, T. K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 (2008).
Kerppola, T. K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286 (2006).
Urban, P., Mignotte, C., Kazmaier, M., Delorme, F. & Pompon, D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272, 19176–19186 (1997).
Ro, D. K., Ehlting, J. & Douglas, C. J. Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from hybrid poplar. Plant Physiol. 130, 1837–1851 (2002).
Bao, H. et al. The developing xylem transcriptome and genome-wide analysis of alternative splicing in Populus trichocarpa (black cottonwood) populations. BMC Genom. 14, 359 (2013).
Karimi, M., Inze, D. & Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Shevchenko, A. et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl Acad. Sci. USA 93, 14440–14445 (1996).
Gehl, C., Waadt, R., Kudla, J., Mendel, R. R. & Hansch, R. New GATEWAY vectors for high throughput analyses of protein–protein interactions by bimolecular fluorescence complementation. Mol. Plant 2, 1051–1058 (2009).
Menke, F. L. et al. Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol. Plant Microbe Interact. 18, 1027–1034 (2005).
Nakagawa, T. et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 (2007).
Zhang, K. et al. Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5, 3274 (2014).
Pradhan Mitra, P. & Loque, D. Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J. Vis. Exp. 87, e51381 (2014).
Zhang, K. et al. An engineered monolignol 4-O-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell 24, 3135–3152 (2012).
Liu, C.-J., Huhman, D., Sumner, L. W. & Dixon, R. A. Regiospecific hydroxylation of isoflavones by cytochrome p450 81E enzymes from Medicago truncatula. Plant J. 36, 471–484 (2003).
Edwards, R. & Kessmann, H. in Molecular Plant Pathology (eds Gurr, S. J. et al.) 45–52 (IRL, Oxford, 1992).
Gou, M. et al. The MYB107 transcription factor positively regulates suberin biosynthesis. Plant Physiol. 173, 1045–1058 (2016).
Acknowledgements
We thank I. Hara-Nishimura at Kyoto University for providing the SP–GFP–HDEL transgenic Arabidopsis seeds. We thank A. Koller at Stony Brook University Proteomics Center for LC–MS analysis of protein complexes. This work was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-SC0012704, specifically through the Physical Biosciences programme of the Chemical Sciences, Geosciences and Biosciences Division (to C.-J.L.). The protein co-immunoprecipitation and sequence analysis was partially supported by the Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Basic Energy Sciences under award no. DE-SC0001090. This research used a confocal microscope of the Center for Functional Nanomaterials, which is a US DOE Office of Science Facility, at Brookhaven National Laboratory under contract no. DE-SC0012704.
Author information
Authors and Affiliations
Contributions
C.-J.L. and M.G. designed the experiments. M.G. and X.R. conducted the experiments. D.W.M processed the proteomic raw data and was responsible for the data deposition. C.-J.L. and M.G. analysed and interpreted the data and wrote the manuscript. All authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–15, Supplementary Tables 1–3 and Supplementary References.
Rights and permissions
About this article
Cite this article
Gou, M., Ran, X., Martin, D.W. et al. The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nature Plants 4, 299–310 (2018). https://doi.org/10.1038/s41477-018-0142-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0142-9
This article is cited by
-
Systematic approaches to C-lignin engineering in Medicago truncatula
Biotechnology for Biofuels and Bioproducts (2023)
-
Directed growth and fusion of membrane-wall microdomains requires CASP-mediated inhibition and displacement of secretory foci
Nature Communications (2023)
-
Dynamics of pectic homogalacturonan in cellular morphogenesis and adhesion, wall integrity sensing and plant development
Nature Plants (2022)
-
Metabolic evidence for distinct pyruvate pools inside plant mitochondria
Nature Plants (2022)
-
Biosynthesis of a novel ganoderic acid by expressing CYP genes from Ganoderma lucidum in Saccharomyces cerevisiae
Applied Microbiology and Biotechnology (2022)