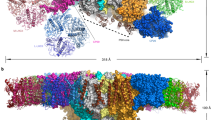
Abstract
Oxygenic photosynthesis produces oxygen and builds a variety of organic compounds, changing the chemistry of the air, the sea and fuelling the food chain on our planet. The photochemical reactions underpinning this process in plants take place in the chloroplast. Chloroplasts evolved ~1.2 billion years ago from an engulfed primordial diazotrophic cyanobacterium, and chlororibosomes are responsible for synthesis of the core proteins driving photochemical reactions. Chlororibosomal activity is spatiotemporally coupled to the synthesis and incorporation of functionally essential co-factors, implying the presence of chloroplast-specific regulatory mechanisms and structural adaptation of the chlororibosome1,2. Despite recent structural information3,4,5,6, some of these aspects remained elusive. To provide new insights into the structural specialities and evolution, we report a comprehensive analysis of the 2.9–3.1 Å resolution electron cryo-microscopy structure of the spinach chlororibosome in complex with its recycling factor and hibernation-promoting factor. The model reveals a prominent channel extending from the exit tunnel to the chlororibosome exterior, structural re-arrangements that lead to increased surface area for translocon binding, and experimental evidence for parallel and convergent evolution of chloro- and mitoribosomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
23 July 2018
In the version of this Article originally published, the name of co-author Annemarie Perez Boerema was coded wrongly, resulting in it being incorrect when exported to citation databases. This has been corrected, though no visible changes will be apparent.
References
Sobotka, R. Making proteins green; biosynthesis of chlorophyll-binding proteins in cyanobacteria. Photosynth. Res. 119, 223–232 (2014).
Barkan, A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155, 1520–1532 (2011).
Ahmed, T., Yin, Z. & Bhushan, S. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci. Rep. 6, 35793 (2016).
Graf, M. et al. Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 45, 2887–2896 (2017).
Bieri, P., Leibundgut, M., Saurer, M., Boehringer, D., & Ban, N. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EM BO J. e201695959
Ahmed, T., Shi, J. & Bhushan, S. Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Nucleic Acids Res. 45, 8581–8595 (2017).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Borovinskaya, M. et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 14, 727–732 (2007).
Weixlbaumer, A. et al. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 14, 498–502 (2007).
Polikanov, Y. S., Blaha, G. M. & Steitz, T. A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918 (2012).
Fleischmann, T. et al. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23, 3137–3155 (2011).
Kramer, G., Boehringer, D., Ban, N. & Bukau, B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 (2009).
Zoschke, R. & Barkan, A. Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc. Natl Acad. Sci. USA 112, 1678–1687 (2015).
Gu, S. Q., Peske, F., Wieden, H. J., Rodnina, M. V. & Wintermeyer, W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9, 566–573 (2003).
Pool, M. R., Stumm, J., Fulga, T. A., Sinning, I. & Dobberstein, B. Distinct modes of signal recognition particle interaction with the ribosome. Science 297, 1345–1348 (2002).
Ullers, R. et al. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 161, 679–684 (2003).
Bornemann, T., Jöckel, J., Rodnina, M. V. & Wintermeyer, W. Signal sequence–independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15, 494–499 (2008).
Bubunenko, M. G., Schmidt, J. & Subramanian, A. R. Protein substitution in chloroplast ribosome evolution: a eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J. Mol. Biol. 240, 28–41 (1994).
Moore, M. J., Soltis, P. S., Bell, C. D., Burleigh, J. G. & Soltis, D. E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl Acad. Sci. USA 107, 4623–4628 (2010).
Weng, M. L., Ruhlman, T. A. & Jansen, R. K. Plastid–nuclear interaction and accelerated coevolution in plastid ribosomal genes in Geraniaceae. Genome Biol. Evol. 8, 1824–1838 (2016).
Jomaa, A., Boehringer, D., Leibundgut, M. & Ban, N. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 7, 10471 (2016)
Breiman, A., Fieulaine, S., Meinnel, T. & Giglione, C. The intriguing realm of protein biogenesis: facing the green co-translational protein maturation networks. Biochim. Biophys. Acta Proteins Proteom. 1864, 531–550 (2016).
Zhang, L., Paakkarinen, V., Suorsa, M. & Aro, E. M. A SecY homologue is involved in chloroplast-encoded D1 protein biogenesis. J. Biol. Chem. 276, 37809–37814 (2001).
Nilsson, R. & Jan van Wijk, K. Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast‐encoded D1 protein; ‘cpSRP54 caught in the act’. FEBS Lett. 524, 127–133 (2002).
Takyar, S., Hickerson, R. P. & Noller, H. F. mRNA helicase activity of the ribosome. Cell 120, 49–58 (2005).
Ban, N. et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 24, 165–169 (2014).
Hood, R. D., Higgins, S. A., Flamholz, A., Nichols, R. J. & Savage, D. F. The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc. Natl Acad. Sci. USA 113, 201524915 (2016).
Sharma, M. R. et al. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl Acad. Sci. USA 104, 19315–19320 (2007).
Galmozzi, C. V., Florencio, F. J. & Muro-Pastor, M. I. The cyanobacterial ribosomal-associated protein LrtA is involved in post-stress survival in Synechocystis sp. PCC 6803. PloS ONE 11, e0159346 (2016).
Matzov, D. et al. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat. Commun. 8, 723 (2017).
Khusainov, I., et al. Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EM BO J. e201696105 (2017).
Van Knippenberg, P. H., Hooykaas, P. J. J. & Van Duin, J. The stoichiometry of E. coli 30S ribosomal protein S1 on in vivo and in vitro polyribosomes. FEBS Lett. 41, 323–326 (1974).
Amunts, A., Brown, A., Toots, J., Scheres, S. H. & Ramakrishnan, V. The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015).
Greber, B. et al. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 (2015).
Puthiyaveetil, S., Ibrahim, I. M. & Allen, J. F. Evolutionary rewiring: a modified prokaryotic gene-regulatory pathway in chloroplasts. Philos. Trans. R. Soc. B 368, 20120260 (2013).
Maier, U.-G. et al. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol. Evol. 5, 2318–2329 (2013).
Allen, J. F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proc. Natl Acad. Sci. 112, 10231–10238 (2015).
Zheng, S., Palovcak, E., Armache, J. P., Cheng, Y. & Agard, D. Anisotropic correction of beam-induced motion for improved single-particle electron cryo-microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 60, 2126–2132 (2004).
Noeske, J. et al. Synergy of streptogramin antibiotics occurs independently of their effects on translation. Antimicrob. Agents Chemother. 58, 5269–5279 (2014).
Minasov, G. et al. 4GFQ: 2.65 Angstrom resolution crystal structure of ribosome recycling factor (frr) from Bacillus anthracis (RCSB Protein Data Bank); http://www.rcsb.org/pdb/explore.do?structureId=4GFQ
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 66, 12–21 (2010).
The PyMOL Molecular Graphics System, Mac v.1.72. (Schrödinger, LLC, 2016).
Pettersen, E. F. et al. UCSF chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Larkin, M. A. et al. ClustalW and ClustalX version 2. Bioinformatics 23, 2947–2948 (2007).
Antczak, M. et al. RNApdbee – a webserver to derive secondary structures from pdb files of knotted and unknotted RNAs. Nucleic Acids Res. 42, 368–372 (2014).
Bernier, C. R. et al. FD169: RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 169, 195–207 (2014).
Ho, B. K. & Gruswitz, F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2006).
Acknowledgements
The data were collected at the UK national electron bio-imaging centre (eBIC, proposal EM15290-2) and the Swedish national cryo-EM facility, funded by the Knut and Alice Wallenberg, and the Family Erling Persson foundations. We thank A. Siebert, D. Clare, M. Carroni and J. Conrad for help with data collection, S. Fleischmann for computing support and A. Petrov for help with the analysis of rRNA. This work was supported by the Swedish Foundation for Strategic Research (Future Leaders Grant FFL15:0325), Ragnar Söderberg Foundation (Fellowship in Medicine M44/16), Swedish Research Council (NT_2015-04107, NT-2013-5901), Raymond & Beverly Sackler Foundation, Swedish e-Science Center FEBS Long-Term Fellowship (SA), Lawski Scholarship (BOF) and Erasmus Mundus Programme (BP).
Author information
Authors and Affiliations
Contributions
A.P.B. and A.A. designed the study. A.P.B., B.P. and A.A. purified chlororibosomes. A.P.B., S.A., B.P. and A.A. prepared cryosamples, optimized conditions and collected data. A.P.B. and S.A. processed data. A.P.B., S.A., B.P., V.T., D.K., B.O.F. and K.W. built the model. A.P.B., S.A. and A.A. wrote the manuscript. All authors discussed and commented on the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–4 and References, and Supplementary Figures 1–10.
Rights and permissions
About this article
Cite this article
Perez Boerema, A., Aibara, S., Paul, B. et al. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nature Plants 4, 212–217 (2018). https://doi.org/10.1038/s41477-018-0129-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0129-6
This article is cited by
-
Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH
Nature Communications (2024)
-
Structure of the actively translating plant 80S ribosome at 2.2 Å resolution
Nature Plants (2023)
-
Modulating co-translational protein folding by rational design and ribosome engineering
Nature Communications (2022)
-
Structure of a mitochondrial ribosome with fragmented rRNA in complex with membrane-targeting elements
Nature Communications (2022)
-
Plastid ribosome protein L5 is essential for post-globular embryo development in Arabidopsis thaliana
Plant Reproduction (2022)