Abstract
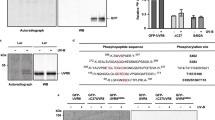
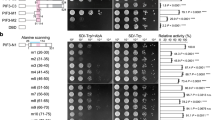
UV RESISTANCE LOCUS 8 (UVR8) is an ultraviolet-B (UVB) radiation photoreceptor that mediates light responses in plants. How plant UVR8 acts in response to UVB light is not well understood. Here, we report the identification and characterization of the Arabidopsis WRKY DNA-BINDING PROTEIN 36 (WRKY36) protein. WRKY36 interacts with UVR8 in yeast and Arabidopsis cells and it promotes hypocotyl elongation by inhibiting HY5 transcription. Inhibition of hypocotyl elongation under UVB requires the inhibition of WRKY36. WRKY36 binds to the W-box motif of the HY5 promoter to inhibit its transcription, while nuclear localized UVR8 directly interacts with WRKY36 to inhibit WRKY36–DNA binding both in vitro and in vivo, leading to the release of inhibition of HY5 transcription. These results indicate that WRKY36 is a negative regulator of HY5 and that UVB represses WRKY36 via UVR8 to promote the transcription of HY5 and photomorphogenesis. The UVR8–WRKY36 interaction in the nucleus represents a novel mechanism of early UVR8 signal transduction in Arabidopsis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Favory, J. J. et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601 (2009).
Jenkins, G. I. Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 60, 407–431 (2009).
Rizzini, L. et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106 (2011).
Jenkins, G. I. Structure and function of the UV-B photoreceptor UVR8. Curr. Opin. Struct. Biol. 29, 52–57 (2014).
Tilbrook, K. et al. The UVR8 UV-B photoreceptor: perception, signaling and response. Arab. Book 11, e0164 (2013).
Kaiserli, E. & Jenkins, G. I. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell. 19, 2662–2673 (2007).
Yi, C. & Deng, X. W. COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell. Biol. 15, 618–625 (2005).
Huang, X., Yang, P., Ouyang, X., Chen, L. & Deng, X. W. Photoactivated UVR8–COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 10, e1004218 (2014).
Cloix, C. et al. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc. Natl Acad. Sci. USA 109, 16366–16370 (2012).
Yin, R., Skvortsova, M. Y., Loubery, S. & Ulm, R. COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc. Natl Acad. Sci. USA 113, E4415–E4422 (2016).
Qian, C. et al. Dual-source nuclear monomers of UV-B light receptor direct photomorphogenesis in Arabidopsis. Mol. Plant 9, 1671–1674 (2016).
Gruber, H. et al. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl Acad. Sci. USA 107, 20132–20137 (2010).
Heijde, M. & Ulm, R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl Acad. Sci. USA 110, 1113–1118 (2013).
Jiao, Y., Lau, O. S. & Deng, X. W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230 (2007).
Ulm, R. et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl Acad. Sci. USA 101, 1397–1402 (2004).
Brown, B. A. et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl Acad. Sci. USA 102, 18225–18230 (2005).
Oravecz, A. et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 18, 1975–1990 (2006).
Brown, B. A. & Jenkins, G. I. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146, 576–588 (2008).
Stracke, R. et al. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell. Environ. 33, 88–103 (2010).
Feher, B. et al. Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 67, 37–48 (2011).
Huang, X. et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 24, 4590–4606 (2012).
Brown, B. A., Headland, L. R. & Jenkins, G. I. UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochem. Photobiol. 85, 1147–1155 (2009).
Osterlund, M. T., Hardtke, C. S., Wei, N. & Deng, X. W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000).
Binkert, M. et al. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell. 26, 4200–4213 (2014).
Liu, H. et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 (2008).
Liu, H. et al. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl Acad. Sci. USA 110, 17582–17587 (2013).
Liu, Y., Li, X., Li, K., Liu, H. & Lin, C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9, e1003861 (2013).
Ma, D. et al. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl Acad. Sci. USA 113, 224–229 (2016).
Pedmale, U. V. et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245 (2016).
Ni, M., Tepperman, J. M. & Quail, P. H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667 (1998).
Leivar, P. & Quail, P. H. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28 (2011).
Castrillo, G. et al. Speeding cis–trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS ONE 6, e21524 (2011).
Eulgem, T., Rushton, P. J., Robatzek, S. & Somssich, I. E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206 (2000).
Riechmann, J. L. et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110 (2000).
Czechowski, T., Bari, R. P., Stitt, M., Scheible, W. R. & Udvardi, M. K. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. Cell. Mol. Biol. 38, 366–379 (2004).
Wu, D. et al. Structural basis of ultraviolet-B perception by UVR8. Nature 484, 214–219 (2012).
Yin, R., Arongaus, A. B., Binkert, M. & Ulm, R. Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell. 27, 202–213 (2015).
Kreuzaler, F., Ragg, H., Fautz, E., Kuhn, D. N. & Hahlbrock, K. UV-induction of chalcone synthase mRNA in cell suspension cultures of Petroselinum hortense. Proc. Natl Acad. Sci. USA 80, 2591–2593 (1983).
Sweere, U. et al. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294, 1108–1111 (2001).
Huq, E. et al. PHYTOCHROME-INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305, 1937–1941 (2004).
Quail, P. H. Photosensory perception and signalling in plant cells: new paradigms? Curr. Opin. Cell. Biol. 14, 180–188 (2002).
Al-Sady, B., Kikis, E. A., Monte, E. & Quail, P. H. Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl Acad. Sci. USA 105, 2232–2237 (2008).
Choi, G. et al. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613 (1999).
Ryu, J. S. et al. Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120, 395–406 (2005).
Fankhauser, C. & Chory, J. Light receptor kinases in plants! Curr. Biol. 9, R123–R126 (1999).
Wang, Q. et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354, 343–347 (2016).
Liu, Q. et al. Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 8, 15234 (2017).
Park, E. et al. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72, 537–546 (2012).
Chen, F. et al. Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell. 26, 1949–1966 (2014).
Acknowledgements
We thank X. Huang, Y. Shi, R. P. Hellens and Q. Hu for materials and technical assistance. We thank G. A. Gomez for copyediting the manuscript. This work is supported in part by the National Key Research and Development Program of China (2017YFA 0503800), National Natural Science Foundation of China (31730009, 31721001, 31670282 and 31670307) and Strategic Priority Research Program ‘Molecular Mechanism of Plant Growth and Development’ (XDPB04).
Author information
Authors and Affiliations
Contributions
Y.Y. and H.L. conceived the project. Y.Y. performed most of the experiments. L.Z., X.G., R.S. and N.S. made some constructs. T.L., X.L. and P.Z. provided materials. K.S. performed some protein expression in E. coli. Y.Y. and H.L. analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary References, Supplementary Figures 1–9 and Supplementary Table 1
Rights and permissions
About this article
Cite this article
Yang, Y., Liang, T., Zhang, L. et al. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nature Plants 4, 98–107 (2018). https://doi.org/10.1038/s41477-017-0099-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-017-0099-0
This article is cited by
-
Phosphorylation of Arabidopsis UVR8 photoreceptor modulates protein interactions and responses to UV-B radiation
Nature Communications (2024)
-
Genome-wide characterization of regulator of chromosome condensation 1 (RCC1) gene family in Artemisia annua L. revealed a conservation evolutionary pattern
BMC Genomics (2023)
-
Light signaling as cellular integrator of multiple environmental cues in plants
Physiology and Molecular Biology of Plants (2023)
-
WRKY transcription factors: evolution, regulation, and functional diversity in plants
Protoplasma (2023)
-
How is UVR8 relevant in plants? New evidence
Plant Growth Regulation (2023)