Abstract
Metabolic associated fatty liver disease (MAFLD) encompasses a broad spectrum of hepatic disorders, including steatosis, nonalcoholic steatohepatitis (NASH) and fibrosis. We demonstrated that phosphoenolpyruvate carboxykinase 1 (PCK1) plays a central role in MAFLD progression. Male mice with liver Pck1 deficiency fed a normal diet displayed hepatic lipid disorder and liver injury, whereas fibrosis and inflammation were aggravated in mice fed a high-fat diet with drinking water containing fructose and glucose (HFCD-HF/G). Forced expression of hepatic PCK1 by adeno-associated virus ameliorated MAFLD in male mice. PCK1 deficiency stimulated lipogenic gene expression and lipid synthesis. Moreover, loss of hepatic PCK1 activated the RhoA/PI3K/AKT pathway by increasing intracellular GTP levels, increasing secretion of platelet-derived growth factor-AA (PDGF-AA), and promoting hepatic stellate cell activation. Treatment with RhoA and AKT inhibitors or gene silencing of RhoA or AKT1 alleviated MAFLD progression in vivo. Hepatic PCK1 deficiency may be important in hepatic steatosis and fibrosis development through paracrine secretion of PDGF-AA in male mice, highlighting a potential therapeutic strategy for MAFLD.
Similar content being viewed by others
Introduction
Metabolic dysfunction-associated fatty liver disease or ‘MAFLD’ (formerly known as ‘NAFLD’) is the most common chronic liver disease worldwide, affecting nearly 25% of US and European adults1. MAFLD is defined by the presence of steatosis in >5% of hepatocytes, regardless of alcohol consumption or other concomitant liver diseases, especially the presence of obesity and T2DM2. It entails a wide spectrum of hepatic clinical conditions, spanning from uncomplicated steatosis to nonalcoholic steatohepatitis (NASH), a more serious form of liver damage hallmarked by irreversible pathological changes such as inflammation, varying degrees of fibrosis, and hepatocellular damage, which is more likely to develop into cirrhosis and hepatocellular carcinoma3. Although multiple parallel insults, including oxidative damage, endoplasmic reticulum stress, and hepatic stellate cell (HSC) activation, have been proposed to explain the pathogenesis of MAFLD, the underlying mechanisms remain unclear4.
In gluconeogenesis, glucose is generated from non-carbohydrate substrates, such as glycerol, lactate, pyruvate, and glucogenic amino acids, mainly in the liver, to maintain glucose levels and energy homeostasis. Phosphoenolpyruvate carboxykinase 1 (PCK1) is the first rate-limiting enzyme in gluconeogenesis and converts oxaloacetate to phosphoenolpyruvate in the cytoplasm5. Our previous studies showed that PCK1 deficiency promotes hepatocellular carcinoma progression by enhancing the hexosamine-biosynthesis pathway6. However, PCK1 regulates not only glucose homeostasis but also lipogenesis by activating sterol-regulatory element-binding proteins7. Patients lacking PCK1 function present diffuse hepatic macrosteatosis concomitant with hypoglycemia and hyperlactacidemia8. Similarly, mice with reduced Pck1 expression develop insulin resistance, hypoglycemia, and hepatic steatosis, indicating the important role of PCK1 in regulating both glucose homeostasis and lipid metabolism9,10. However, the precise role of PCK1 in MAFLD progression is not well-understood.
The phosphoinositide 3-kinase/protein kinase B (PI3K/ATK) pathway plays a critical role in regulating cell growth and metabolism. This pathway is activated in response to insulin, growth factors, energy, and cytokines and, in turn, regulates key metabolic processes such as glucose and lipid metabolism and protein synthesis11. AKT promotes de novo lipogenesis primarily by activating sterol regulatory element-binding protein12. PI3K/AKT dysregulation leads to numerous pathological metabolic conditions, including obesity and type 2 diabetes13. MAFLD is characterized by dysregulated glucose and lipid metabolism in the liver. Although the PI3K/AKT pathway is a key regulator for sensing metabolic stress, its exact role in MAFLD progression is unclear14,15.
In this study, we explored the role of Pck1 in a mouse model. We determined the molecular mechanisms underlying disordered lipid metabolism, inflammation, and fibrosis induced by Pck1 depletion. We also delineated the functional importance of the PI3K/AKT pathway and paracrine secretion of PDGF-AA as its effectors in steatohepatitis, providing a potential therapeutic strategy for treating MAFLD.
Results
PCK1 is downregulated in patients with NASH and mouse models of MAFLD
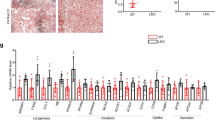
To determine whether PCK1 is involved in MAFLD, we first examined hepatic gene expression in a published transcriptome dataset [Gene Expression Omnibus (GEO): GSE126848] containing samples from 14 healthy participants, 12 patients with obesity, 15 patients with NAFLD, and 16 patients with NASH16. Bioinformatics analysis showed that 32 genes were markedly changed in obesity, NAFLD, and NASH; 12 genes were considerably downregulated and 20 genes were upregulated (Supplementary Fig. 1a–c). Notably, PCK1 was gradually reduced in patients with obesity, NAFLD, and NASH (Fig. 1a, b). Downregulation of PCK1 mRNA was also observed in a similar dataset (GSE89632) (Fig. 1b). Moreover, immunohistochemistry (IHC) assays showed that hepatic PCK1 protein levels were significantly lower in patients with NASH than in healthy participants (Fig. 1c). Similarly, PCK1 mRNA and protein levels decreased in the liver of mice fed a high-fat diet with drinking water containing fructose and glucose (HFCD-HF/G) for 24 weeks (Fig. 1d, e).
a Genes downregulated in patients with health (n = 14), obesity (n = 12), NAFLD (n = 15), and NASH (n = 16) from GSE126848 dataset. b Relative PCK1 mRNA levels of health (n = 14), obesity (n = 12), NAFLD (n = 15), and NASH (n = 16) in GSE126848 and of health (n = 19), and NASH (n = 24) in GSE89632 datasets. The box plots show the medians (middle line) and the first and third quartiles (boxes), whereas the whiskers show 1.5× the IQR above and below the box. Unpaired, two-sided Mann–Whitney U test P values are depicted in the plots, and the significant P value cutoff was set at 0.05. c PCK1 expression in normal individuals and patients with NASH and semi-quantitative analyses of immunohistochemistry (IHC) data (health, n = 10; NASH, n = 36). Scale bars: 50 µm. d, e mRNA (d) and protein (e) levels of PCK1 in the livers of WT mice fed with chow diet or HFCD-HF/G (n = 6). n was the number of biologically independent mice. The samples were derived from the same experiment and the blots were processed in parallel. f, g PCK1 mRNA (f) and protein (g) levels in MIHA cells treated with BSA or PA-BSA. h Relative levels of indicated genes in MIHA cells treated with 0.2 mM PA. i, j Representative ATF3 expression in normal individuals and patients with NASH (i) and semiquantitative analyses of IHC data (j) (health, n = 10; NASH, n = 36). Scale bars: 50 µm. k Chromatin immunoprecipitation assays were performed in MIHA cells with or without PA treatment using an antibody against ATF3, IgG, or H3. l Protein levels of PCK1 in MIHA cells infected with either shControl or shATF3 treated with 0.2 mM PA. The samples were derived from the same experiment and the blots were processed in parallel. m Correlation analysis of ATF3 mRNA level with PCK1 in human NAFLD/NASH liver samples (GSE135251, n = 206). For f and h, n = 3. Data are expressed as the mean ± SEM; n.s., not significant. p values obtained via two-tailed unpaired Student’s t tests, one-way analysis of variance with Tukey’s post hoc test, or non-parametric Spearman’s test. Source data are provided as a Source Data file.
Next, palmitic acid (PA) was used to mimic the liver steatosis of patients with MAFLD in vitro17. Cell growth was assessed in a CCK8 assay after treatment with different concentrations of PA (Supplementary Fig. 1d). Interestingly, the PCK1 mRNA and protein levels were downregulated in a dose-dependent manner during 24 h PA stimulation (Fig. 1f, g), suggesting that the transcription of PCK1 was inhibited in response to lipid overload. We screened several known regulators of PCK1 (Supplementary Fig. 1e, f) and found that activating transcription factor 3 (ATF3), a transcriptional repressor of PCK118, was upregulated upon PA stimulation (Fig. 1h). Similarly, ATF3 expression was remarkably upregulated in liver samples derived from patients with NASH and MAFLD model mice (Fig. 1i, j, Supplementary Fig. 1g, h). Chromatin immunoprecipitation assays revealed that the binding of ATF3 to the PCK1 promoter increased following PA administration (Fig. 1k). ATF3 knockdown considerably enhanced PCK1 promoter activity and restored PCK1 expression in human hepatocytes treated with PA (Fig. 1l), while overexpression of ATF3 played a opposite effects on PCK1 promoter activity (Supplementary Fig. 1i, j). Furthermore, correlation analysis revealed that ATF3 was negatively correlated with the PCK1 mRNA level based on the GEO database (GEO: GSE135251) (Fig. 1m). These results indicate that increased lipid intake led to the upregulation of the repressor ATF3, impairing PCK1 transcription in patients with NASH and mouse models.
L-KO mice exhibit a distinct hepatic steatosis phenotype
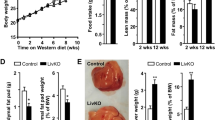
To explore the role of Pck1 in fatty liver disease, wild-type (WT) and liver-specific Pck1-knockout mice (L-KO) mice were fed a chow diet for 24 weeks (Fig. 2a). Hepatic-specific depletion of PCK1 was confirmed by performing immunoblotting (Fig. 2b). Starting at 16 weeks, L-KO mice showed an increased body weight compared with WT mice, however, the results of the glucose tolerance test (GTT) and insulin tolerance test (ITT) did not significantly differ (Fig. 2c). Moreover, increased liver weight was observed in L-KO mice (Fig. 2d). Alanine transaminase (ALT) and aspartate transaminase (AST) levels were higher in L-KO mice, indicating liver injury (Fig. 2e). In addition, total triglyceride (TG), total cholesterol (TC), and free fatty acids (FFAs) in the liver tissues and serum were elevated in L-KO mice compared to those in WT mice (Fig. 2f, g). Histochemistry and enzyme-linked immunosorbent assay (ELISA) showed that L-KO mice had prominent hepatic steatosis, increased inflammatory infiltration, and high levels of TNF-α, whereas hepatic fibrosis was not observed (Fig. 2h–j). Additionally, PCK1 deficiency significantly increased the mRNA levels of genes related to fatty acid transport and inflammation, whereas there were no significant changes in fibrosis-related genes in L-KO mice fed the chow diet (Fig. 2k). These data suggest that L-KO mice exhibited a distinct hepatic steatosis phenotype and liver injury even when fed normal chow.
a Schematic diagram of the mouse model fed the chow diet, n = 10/group. b PCK1 protein expression in WT and L-KO mouse intestine, liver, spleen, kidney, white adipose, and brown adipose confirmed by immunoblotting. The samples were derived from the same experiment and the blots were processed in parallel. This experiment was repeated for three times with similar results. c Body weight, glucose tolerance test (GTT), and insulin tolerance test (ITT) were measured in WT and L-KO mice (n = 10). d Liver weight of WT and L-KO mice (n = 10). e–g Determination of alanine aminotransferase (ALT), aspartate aminotransferase (AST), total triglycerides (TG), total cholesterol (TC), and free fatty acid (FFA) levels in the serum or liver tissues (n = 10). h Paraffin-embedded liver sections were stained with hematoxylin and eosin (H&E), Sirius red, and F4/80. Frozen sections stained with Oil Red O. Scale bars: 50 µm. i Quantification of liver sections of WT and L-KO mice fed the chow diet (n = 10). j Levels of TNF-α and IL-6 in the liver tissues (n = 10). k Quantitative PCR analysis of liver mRNA expression (n = 10). For immunoblotting, the samples were derived from the same experiment and the blots were processed in parallel. n was the number of biologically independent mice. Data are expressed as the mean ± SEM; n.s., not significant. p values obtained via two-tailed unpaired Student’s t tests. Source data are provided as a Source Data file.
Hepatic loss of Pck1 promotes inflammation and fibrogenesis in MAFLD mice
To explore whether an unhealthy diet could exacerbate pathologic changes in L-KO mice, WT and L-KO mice were fed HFCD-HF/G (Fig. 3a)19,20. Starting at 4 weeks, L-KO mice showed significant weight gain (Fig. 3b). The GTT and ITT showed that L-KO mice developed a more severe form of glucose intolerance and insulin resistance compared with those in WT mice (Supplementary Fig. 2a, b). L-KO mice had heavier livers compared with WT mice (Fig. 3c), although there was no significant difference in the liver weight ratio (Supplementary Fig. 2c). Insulin, AST, ALT, TC, TG, and FFAs increased in the serum and liver homogenates of L-KO mice, suggesting more serious liver injury and lipid metabolism disorder (Fig. 3d, e, Supplementary Fig. 2d, e). Analyses of L-KO liver sections revealed increased fat droplets, more severe fibrosis, and greater macrophage infiltration (Fig. 3f, Supplementary Fig. 2f). Furthermore, L-KO mice exhibited higher NAFLD activity scores (NAS score) and higher TNF-α and IL-6 levels (Fig. 3g, h). In addition, the expression of inflammatory factors, lipogenic enzymes, and fibrogenesis-associated genes was upregulated in L-KO mice (Supplementary Fig. 2g). In summary, mice lacking hepatic Pck1 showed substantial liver inflammation and fibrosis after being fed the HFCD-HF/G.
a Schematic diagram of mouse model fed the HFCD-HF/G. b, c Body weight (n = 11 per group) and liver weight (n = 8 per group) were measured in WT and L-KO mice. d, e Serum levels of insulin, alanine aminotransferase (ALT), and aspartate aminotransferase (AST) were measured (n = 8). f Paraffin-embedded liver sections were stained with hematoxylin and eosin (H&E), Sirius red, α-SMA immunostaining, and F4/80 immunostaining. Frozen sections stained with Oil Red O. Scale bars: 50 µm. g NAFLD activity scores (NAS) for each group (n = 8). h Levels of TNF-α and IL-6 in the liver tissues detected using enzyme-linked immunosorbent assay (ELISA) (n = 8). i Schematic showing the administration protocol for AAV8-TBG-control or AAV8-TBG-Pck1 in WT and L-KO mice for experiments shown in j–l, n = 6/group. j Liver weight and body weight. k Paraffin-embedded liver sections were stained with H&E, Sirius red staining and Oil Red O staining. Scale bars: 50 µm. l Quantifications of Sirius red staining, Oil Red O staining, and NAS. Data are expressed as the mean ± SEM; n.s., not significant. p values obtained via two-tailed unpaired Student’s t tests. Source data are provided as a Source Data file.
Besides, we used a genetic model with hepatic deficiency in phosphatase and tensin homolog (PTEN) to induce MAFLD. We generated a mouse model with biallelic deletion of Pck1 and Pten in the liver (cPtenf/fPck1f/f) (Supplementary Fig. 3a, b). At 6 months, cPtenf/fPck1f/f mice had higher liver/body weight ratios compared with cPtenf/f mice (Supplementary Fig. 3c). ALT, AST, IL-6, TNF-a, TC, TG, and FFAs levels significantly increased in the serum and liver tissues of cPtenf/fPck1f/f mice, suggesting more serious liver injury and lipid metabolism disorder in cPtenf/fPck1f/f mice (Supplementary Fig. 3d, e). Histological analysis of cPtenf/fPck1f/f liver sections revealed increased fat droplets, more severe fibrosis, and greater macrophage infiltration (Supplementary Fig. 3f, g). In agreement with increased inflammation and fibrosis, elevated expression levels of genes related to inflammation and fibrosis were observed in cPtenf/fPck1f/f mice (Supplementary Fig. 3h). In summary, hepatic Pck1 depletion showed substantial liver steatosis, inflammation and fibrosis in PTEN-null livers.
AAV-mediated restoration of hepatic PCK1 alleviates the MAFLD phenotype in Pck1-deficient mice
We then investigated whether adeno-associated virus (AAV)-based Pck1 replacement therapy could reverse ongoing liver derangement, which is typically observed in patients with MAFLD. After 10 weeks of chow diet or HFCD-HF/G feeding, WT and L-KO mice were injected through the tail vein with AAV serotype 8 (AAV8) vector expressing Pck1 under the control of a liver-specific promoter (thyroxine-binding globulin, TBG), AAV8-TBG-Pck1 or AAV8-TBG-control (Fig. 3i). The expression of Pck1 mRNA and PCK1 protein levels was identified (Supplementary Fig. 4a, b). Interestingly, mice with PCK1 re-expression showed a lower liver weight, body weight, serum liver enzymes, and serum lipid contents (Fig. 3j, Supplementary Fig. 4c, d). Moreover, lipid deposition, inflammation, and fibrosis significantly improved in L-KO mice injected with AAV8-TBG-Pck1 (Fig. 3k-l). Hepatic gene expression analyses indicated that the expression of genes involved in inflammation and liver fibrosis was greatly attenuated by PCK1 restoration in L-KO mice (Supplementary Fig. 4e). Overall, these data support that forced PCK1 expression in the liver protects against MAFLD in mice.
Transcriptomic and metabolomics analyses confirmed that the loss of Pck1 promotes hepatic lipid accumulation
To comprehensively investigate the role of Pck1 deficiency in MAFLD development, we performed RNA-seq analysis of liver samples from L-KO and WT mice fed the normal chow diet or HFCD-HF/G for 24 weeks. Gene Ontology analysis indicated that lipid metabolic processes were remarkably upregulated in L-KO mice fed the HFCD-HF/G (Fig. 4a). The volcano plot showed that genes involved in fatty acid uptake, such as solute carrier family 27 member 1 (Slc27a1) and fatty acid translocase (Cd36), and lipid droplet synthesis, such as cell death-inducing DFFA like effector C (Cidec) and cell death-inducing DFFA like effector A (Cidea), were upregulated in response to the HFCD-HF/G (Fig. 4b). Gene Set Enrichment Analysis (GSEA) revealed that the PPAR signaling pathway was prominently upregulated in L-KO mice fed either diet (Fig. 4c, Supplementary Fig. 5a, b). Several genes selected from the dataset were independently validated by quantitative polymerase chain reaction (qPCR) and immunoblotting and found to be significantly overexpressed in L-KO mice (Fig. 4d, e). Furthermore, genes involved in the glycerol 3-phosphate (G3P) pathway were upregulated in L-KO mice (Fig. 4f). Metabolomics analysis showed that compared with WT mice fed the HFCD-HF/G, L-KO mice had significantly higher G3P and PA levels (Fig. 4g, h). As G3P is a substrate for TG synthesis and PA is a key intermediate metabolite in de novo lipogenesis, Pck1 ablation may promote substrate accumulation for lipid synthesis.
RNA sequencing was performed on the livers of WT (n = 4) and L-KO (n = 5) mice fed the HFCD-HF/G. a Gene Ontology analysis of all significantly changed genes in top 10 biological processes. b Volcano plot representation of significantly up- and downregulated genes. c Gene Set Enrichment Analysis plot (left) of enrichment in “PPAR signaling pathway” signature; heatmap (right) of significantly upregulated PPAR target genes. d, e Quantitative PCR (d) and immunoblot (e) analysis of indicated genes or protein expression in mouse liver tissues. f Relative mRNA expression of key genes in G3P pathway (n = 8). g Upregulated metabolites detected by untargeted metabolomics (n = 6). h Relative level of G3P and PA in mouse liver tissues (n = 6). Data are expressed as the mean ± SEM. p values obtained via two-tailed unpaired Student’s t tests. Source data are provided as a Source Data file.
To further examine the function of PCK1 in steatosis in vitro, we overexpressed (PCK1-OE) using the AdEasy adenoviral vector system and knocked out PCK1 (PCK1-KO) using the CRISPR-Cas9 system in human hepatocytes (Supplementary Fig. 5c, d). We found that PCK1-OE attenuated the accumulation of lipid droplets, whereas PCK1-KO facilitated lipid accumulation (Supplementary Fig. 5e, f). Collectively, these results suggest that hepatic Pck1 deficiency leads to lipid accumulation by promoting the expression of lipogenic genes and accumulation of substrates related to lipid synthesis (Supplementary Fig. 5g).
Hepatic Pck1 deficiency leads to HSC activation via PI3K/AKT pathway
RNA-seq analysis indicated that the PI3K/AKT pathway was specifically activated in L-KO mice fed the HFCD-HF/G (Fig. 5a, b). Immunoblotting revealed p-AKT (S473) and p-AKT (T308), which are two activated forms of AKT, and downstream c-MYC were significantly upregulated in both the livers and primary hepatocytes of L-KO mice fed the HFCD-HF/G (Fig. 5c, d). qPCR confirmed that genes related to the PI3K/AKT pathway were highly expressed in L-KO mice (Supplementary Fig. 6a). Similarly, p-AKT (S473 and T308) significantly decreased in human PCK1-OE cells but increased in PCK1-KO cells after 0.2 mM PA treatment (Fig. 5e, f).
a Pathway enrichment analysis of significantly upregulated genes in L-KO mice. b Gene Set Enrichment Analysis (GSEA) plot of enrichment in PI3K/AKT pathway. c–f Immunoblot analysis of AKT and p-AKT (S473 or T308) in mouse liver tissues (c), primary hepatocytes from HFCD-HF/G feeding mice (d), PCK1-OE (e), and PCK1-KO (f) MIHA cells with or without 0.2 mM palmitic acid (PA) treatment. The samples were derived from the same experiment and the blots were processed in parallel. g Schematic flow chart of co-culture models. h, i Quantitative PCR analysis of fibrosis-related genes in LX-2 cells co-cultured with PCK1-KO (h) or PCK1-OE (i) MIHA cells. j, k Western blotting of fibrosis-related protein in liver tissues (j) or primary HSCs from HFCD-HF/G feeding mice (k) (n = 3) l, m Relative mRNA expression (l) and immunofluorescence images (m) of ACTA2/α-SMA, COL1A1, and COL3A1 in LX-2 cells co-cultured with PCK1-KO MIHA cells treated with AKT inhibitor MK2206 (10 μM). Scale bars: 25 µm. Data are expressed as the mean ± SEM. p values obtained via two-tailed unpaired Student’s t tests or one-way ANOVA with Tukey’s post hoc test. Source data are provided as a Source Data file.
To clarify the role of PI3K/AKT pathway activation, transcriptome data were further analyzed. Interestingly, Col1a1, Col3a1, and Lama2, which are primary components of the extracellular matrix (ECM), were upregulated, as shown in the heat map of the PI3K/AKT pathway (Supplementary Fig. 6b). Moreover, GSEA analysis revealed that ECM-receptor interaction was upregulated in L-KO mice (Supplementary Fig. 6c). Because ECM deposition is typically considered as the key event underlying liver fibrosis, we predicted that the activation of the PI3K/AKT pathway promotes fibrosis in L-KO mice. HSCs are major ECM secretors; thus, we performed co-culture assays with human hepatocyte (MIHA) and human hepatic stellate cell lines (LX-2) (Fig. 5g). Interestingly, the mRNA levels of ACTA2 (α-SMA, an HSC activation marker), COL1A1, and COL3A1 increased in LX-2 cells co-cultured with PCK1-KO cells but decreased in LX-2 cells co-cultured with PCK1-OE cells (Fig. 5h, i). Similarly, COL1A1, COL3A1, and α-SMA expression increased in the liver tissues and primary HSCs of L-KO mice (Fig. 5j, k), which was confirmed in IHC analysis of COL3A1 (Supplementary Fig. 6d). However, these increases were partially reversed by MK2206, an AKT inhibitor (Fig. 5l, m). Collectively, these data suggest that the loss of PCK1 in hepatocytes induces HSC activation and ECM formation by activating the PI3K/AKT pathway.
Paracrine PDGF-AA from hepatocytes promotes HSC activation
Hepatocytes elicit several fibrogenic actions in a paracrine manner to promote HSC activation21. Thus, PCK1-mediated hepatic fibrosis may be involved in paracrine disorders. To test this hypothesis, several pro-fibrotic factors were screened; Pdgfa was found to be significantly elevated in the liver tissues of L-KO mice (Fig. 6a). Bioinformatics analysis confirmed that PDGFA was significantly increased in patients with NAFLD and NASH (Fig. 6b). Pdgfa encodes a dimer disulfide-linked polypeptide (PDGF-AA), and the chronic elevation of PDGF-AA in the mouse liver induces fibrosis22. Immunoblotting and ELISA revealed increased PDGF-AA expression in the liver tissues, primary hepatocytes, and plasma of L-KO mice (Fig. 6c–f). Moreover, the PDGF-AA concentration markedly increased in the culture medium of PCK1-KO cells but decreased in that of PCK1-OE cells treated with 0.2 mM PA (Fig. 6g, h). Correspondingly, platelet-derived growth factor receptor alpha (PDGFRA), which encodes the PDGF-AA receptor, increased in LX-2 cells co-cultured with PCK1-KO cells but decreased in LX-2 cells co-cultured with PCK1-OE cells (Fig. 6i, j). To determine whether the pro-fibrogenic effect was mediated by PDGF-AA secretion, we treated the cells with a neutralizing antibody against PDGF-AA. As expected, the increases in α-SMA, COL1A1, and COL3A1 in LX-2 cells co-cultured with PCK1-KO cells were reversed by anti-PDGF-AA treatment (Fig. 6k).
a Expression levels of genes related to fibrogenesis (n = 8 for each group). b Relative PDGFA mRNA levels of health (n = 14), obesity (n = 12), NAFLD (n = 15), and NASH (n = 16) in GSE126848 and of health (n = 19), and NASH (n = 24) in GSE89632 datasets. The box plots show the medians (middle line) and the first and third quartiles (boxes), whereas the whiskers show 1.5× the IQR above and below the box. Unpaired, two-sided Mann–Whitney U test P values are depicted in the plots, and the significant P value cutoff was set at 0.05. c, d PDGF-AA protein levels in primary hepatocytes (c) or liver tissues (d) detected by western blotting. The samples were derived from the same experiment and the blots were processed in parallel. e, f PDGF-AA levels in liver tissues (e) or serum (f) were detected using enzyme-linked immunosorbent assay (ELISA) (n = 8). g, h Secreted PDGF-AA levels in conditioned medium with PCK1-KO (g) or PCK1-OE (h) MIHA cells treated with 0.2 mM palmitic acid (PA). i, j mRNA levels of PDGFRA in cell lysate of LX-2 cells co-cultured with PCK1-KO (i) or PCK1-OE (j) MIHA cells treated with PA. k Protein level in LX-2 cells co-cultured with PCK1-KO MIHA cells containing nonspecific rabbit IgG or a PDGF-AA blocking antibody. I Immunohistochemistry (IHC) analysis of PCK1, p-AKT (S473), and PDGF-AA in mouse liver sections (from serial sections). Scale bars: 50 µm. m, n Levels of PDGF-AA (m) or PDGFA (n) in conditioned medium or cell lysate of PCK1-KO MIHA cells treated with AKT inhibitor MK2206 (10 μM). o Protein levels in PCK1-KO MIHA cells treated with AKT inhibitor MK2206 (10 μM). The samples were derived from the same experiment and the blots were processed in parallel. For g, h, i, j, m and n, n = 3. Data are expressed as the mean ± SEM; n.s., not significant. p values obtained via two-tailed unpaired Student’s t tests or one-way ANOVA with Tukey’s post hoc test. Source data are provided as a Source Data file.
A review of the transcriptome data showed that Pdgfa appeared in the heat map of the PI3K/AKT pathway (Supplementary Fig. 6b). The IHC results showed that p-AKT (S473) was positively correlated with PDGF-AA (Fig. 6l). The AKT inhibitor MK2206 significantly blocked the increase in PDGFA levels in the supernatants or cells lysates of PCK1-KO cells (Fig. 6m–o). Taken together, these data confirm that PCK1 deficiency promoted PDGF-AA expression through the PI3K/AKT pathway and activated HSCs through hepatocyte-HSC crosstalk.
PCK1 deficiency promotes the activation of the PI3K/AKT/PDGF-AA axis by activating RhoA signaling in hepatocytes
Rho GTPases, which cycle between active GTP-bound and inactive GDP-bound conformations, activate the PI3K/AKT pathway23,24,25. Considering that PCK1 catalyzes the conversion of oxaloacetate to phosphoenolpyruvate, consuming GTP to generate GDP, we predicted that PCK1 deficiency alters intracellular GTP homeostasis. To test this hypothesis, high-performance liquid chromatography (HPLC) analysis was conducted to detect the intracellular levels of GTP. Interestingly, intracellular GTP levels decreased in PCK1-OE cells (Supplementary Fig. 7a) but increased in PCK1-KO cells (Supplementary Fig. 7b). Considering that Rho GTPases are activated when combined with GTP26, we examined the proteins levels of several Rho GTPases in the mouse liver tissues and found that GTP-bound RhoA significantly increased and inactivated RhoA, p-RhoA (S188), decreased in L-KO mice (Fig. 7a–c). Similar results were observed in the primary hepatocytes of mice fed the HFCD-HF/G (Supplementary Fig. 7c). Consistently, after PA treatment, the levels of GTP-bound RhoA decreased and p-RhoA (S188) expression increased in PCK1-OE cells, whereas the opposite results were observed in PCK1-KO cells (Fig. 7d–g). Moreover, we found that the addition of 5’-GTP, 2Na+ (salt of guanosine triphosphate) activated the RhoA/PI3K/AKT pathway in primary hepatocytes (Supplementary Fig. 7d). However, inhibition of de novo guanine nucleotide synthesis by mycophenolic acid (MPA) robustly inhibited the RhoA/PI3K/AKT pathway by decreasing the extent of phosphorylated AKT (Supplementary Fig. 7e). Next, RhoA inhibitor (Rhosin) or shRhoA was used to determine whether PI3K/AKT activation depends on RhoA. Immunoblotting, ELISA, and qPCR assays showed that both Rhosin and shRhoA blocked the increase in the activated forms of AKT and PDGF-AA in the PCK1-KO cell lysate and supernatant, as well as ACTA2, COL1A1, and COL3A1 expression in LX-2 cells co-cultured with PCK1-KO hepatocytes (Fig. 7h–j, Supplementary Fig. 7f–h). To further evaluate the involvement of RhoA in HSC activation, we isolated primary hepatocytes from HFCD-HF/G-fed WT or L-KO mice; treated the cells with MK2206, Rhosin, or dimethyl sulfoxide vehicle; and then co-cultured the cells with primary HSCs from chow-fed WT mice. The activation of co-cultured HSCs was partially eliminated by the inhibition of AKT or RhoA in primary hepatocytes (Supplementary Fig. 7i). Moreover, PCK1 and p-RhoA (S188) were downregulated in samples from patients with NASH, whereas p-AKT (S473) and PDGF-AA levels were upregulated (Fig. 7k, l). These data indicate that PCK1 ablation stimulated the PI3K/AKT/PDGF-AA axis by activating RhoA.
a Immunoblotting analysis of indicated proteins in mouse liver tissues. The samples were derived from the same experiment and the blots were processed in parallel. b Immunohistochemistry (IHC) analysis of p-RhoA (S188) in mouse liver tissues. Scale bars: 50 µm. c–e Relative levels of active RhoA were measured using G-LISA colorimetric RhoA activation assay in mouse liver tissues (c) (n = 8), PCK1-KO (d) and PCK1-OE (e) MIHA cells treated with 0.2 mM palmitic acid (PA). f, g Immunoblots of p-RhoA (S188) and RhoA in PCK1-KO (f) and PCK1-OE (g) MIHA cells with or without 0.2 mM PA treatment. The samples were derived from the same experiment and the blots were processed in parallel. h Expression of indicated proteins in PCK1-KO MIHA cells after addition of Rhosin (30 µM). i Levels of PDGF-AA in the supernatant of PCK1-KO MIHA cells treated with Rhosin (30 µM). j Relative mRNA expression of ACTA2, COL1A1, and COL3A1 in LX-2 cells co-cultured with PCK1-KO MIHA cells treated with Rhosin (30 µM). k IHC analysis of PCK1, p-RhoA (S188), p-AKT (S473), and PDGF-AA in normal individuals and patients with NASH (from serial sections). Scale bars: 50 µm. l Semi-quantitative analyses of immunohistochemistry data of health and NASH human tissues for indicated proteins (health, n = 10, NASH, n = 10). The cell culture experiments were repeated for three times independently with similar results. For d, e, i and j, n = 3. Data are expressed as the mean ± SEM. p values obtained via one-way ANOVA with Tukey’s post hoc test. Source data are provided as a Source Data file.
Genetic or pharmacological disruption of RhoA and AKT1 reduced progressive liver fibrosis in vivo
To explore whether blocking RhoA/PI3K/AKT could rescue the MAFLD phenotype in L-KO mice, the genetic and pharmacological disruption of RhoA and AKT1 was performed in vivo (Fig. 8a). Pharmacological inhibition of AKT1 or RhoA led to improved glucose intolerance (Supplementary Fig. 8a) and insulin resistance (Supplementary Fig. 8b). The increase in liver weight was also prevented (Fig. 8b), whereas the body weight decreased only in the MK2206 treatment group (Supplementary Fig. 8c). Additionally, Rhosin or MK2206 administration attenuated the levels of inflammatory factors (e.g. AST, ALT, IL-6, and TNF-α) as well as TG and FFA levels, in the serum and liver tissues (Fig. 8c, Supplementary Fig. 8d, e). Similarly, histochemistry revealed reduced liver steatosis, inflammation, and fibrosis in Rhosin- or MK2206-treated mice (Fig. 8d, Supplementary Fig. 8f). Additionally, α-SMA, COL1A1, COL3A1, PDGF-AA, and p-AKT (S473, T308) expression and GTP-bound RhoA levels decreased, whereas p-RhoA (S188) expression increased in the treatment group (Supplementary Fig. 8g, h). MK2206 or Rhosin treatment also reduced the expression of genes related to inflammation and fibrosis (Supplementary Fig. 8i). These results indicate that the pharmacological inhibition of AKT or RhoA can alleviate the clinical phenotypes of MAFLD.
L-KO mice were fed the HFCD-HF/G for 24 weeks, and therapeutic treatments were initiated at different times. a Schematic diagram of in vivo pharmacological inhibition (b–d) and pSECC lentivirus-mediated silencing (e–g) of AKT1 or RhoA. b, c Liver weight (b), and serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), TNF-α, and IL-6 (c) (DMSO, n = 6; MK2206, n = 6; Rhosin, n = 5). d Paraffin-embedded liver sections were stained with hematoxylin and eosin, Sirius Red, or immunostained for F4/80, COL3A1, and α-SMA. Frozen sections were stained with Oil Red O. Scale bars: 50 µm. e Quantification of body weight and liver weight in pSECC-sgAkt1 and pSECC-sgRhoA L-KO mice (n = 6). f Plasma levels of total triglycerides (TG), total cholesterol (TC), and free fatty acids (FFA) (n = 6). g NAFLD activity scores of liver sections (n = 6). h Model depicting the critical role of PCK1 in controlling MAFLD progression. n was the number of biologically independent mice. Data are expressed as the mean ± SEM; n.s., not significant. p values obtained via one-way ANOVA with Tukey’s post hoc test. Source data are provided as a Source Data file.
Next, we used pSECC, a lentiviral-based system that combines the CRISPR system and Cre recombinase, to silence RhoA or AKT1 in L-KO mice27. In agreement with the results of the experiments described above, AKT or RhoA depletion in vivo partially prevented and liver weight and body weight gain (Fig. 8e). Moreover, serum AST, ALT, TC, TG, and FFA levels significantly decreased in sgAkt1- or sgRhoA-treated mice compared with those in sgCtrl-treated L-KO mice (Fig. 8f, Supplementary Fig. 9a). Histological analysis revealed that the NAS score, hepatic fibrosis, and F4/80+ macrophage counts decreased in sgAkt1 and sgRhoA mice, indicating the attenuation of the MAFLD phenotypes (Fig. 8g, Supplementary Fig. 9b, c). Consistently, genes or proteins involved in hepatic inflammation and fibrosis were significantly downregulated in the livers of sgAkt1 and sgRhoA mice, accompanied by the reduced expression of PDGF-AA (Supplementary Fig. 9d, e). These results indicate that the genetic inhibition of AKT or RhoA can alleviate the clinical phenotypes of MAFLD.
Discussion
We found that the hepatic gluconeogenic enzyme PCK1 plays an important role in MAFLD progression. The expression of PCK1 was diminished in the livers of patients or mice with MAFLD. Moreover, the deletion of PCK1 significantly exacerbated hepatic steatosis, fibrosis, and inflammation in mouse models fed the HFCD-HF/G. Mechanistically, loss of PCK1 not only promotes steatosis by enhancing lipid deposition, but also induces fibrosis through HSC activation via paracrine secretion of PDGF-AA, thus promoting MAFLD progression (Fig. 8h).
Abnormal lipid metabolism is characteristic of MAFLD. Previous studies predicted that altered lipid homeostasis was caused by abnormal expression of genes related to lipid metabolism28. However, recent studies demonstrated that disruption of gluconeogenesis also leads to abnormal lipid metabolism. A deficiency of fructose-1,6-bisphosphatase 1 and glucose-6-phosphatase catalytic subunit, which are key enzymes in gluconeogenesis, results in severe hepatic steatosis and hypoglycemia, indicating that the suppression of gluconeogenesis also disrupts lipid homeostasis17,29. As the first rate-limiting enzyme in gluconeogenesis, it is currently unclear whether PCK1 plays a critical role in MAFLD development. We identified a robust decrease in PCK1 expression in the livers of MAFLD mice and patients with NAFLD/NASH, causing severe hepatic steatosis and confirming that disordered hepatic gluconeogenesis affects lipid homeostasis.
Previous reports showed that PCK1 expression is increased in several obesity/diabetes mouse models, such as ZDF rats and ob/ob and db/db mice, and the disease progression of MAFLD is positively correlated with obesity and type 2 diabetes mellitus30,31,32. Interestingly, we found that PCK1 expression was downregulated in a diet-induced murine model. This discrepancy may be related to differences in the animal models used in different studies. Widely used rodent models for genetic forms of obesity and diabetes, such as ob/ob and db/db mice, exhibit increased plasma glucocorticoids, which may drive PCK1 expression32,33. Another explanation is that the high-fat diet supplemented with high fructose/glucose in drinking water might suppress PCK1 expression34. Under a high-fat diet, PA decreased PCK1 expression via SIRT3 inhibition35. Additionally, acetylation, ubiquitination, and phosphorylation modulate PCK1 expression36. A high glucose level was reported to destabilize PCK1 by stimulating its acetylation, thus promoting its ubiquitination and subsequent degradation37. In this study, we found that ATF3, a member of the basic leucine zipper family of transcription factors38, transcriptionally repressed Pck1 upon PA overload in vitro and in the mouse model. This result agrees with those of previous studies suggesting that ATF3 is upregulated in patients with NAFLD and murine models and inhibits the expression of PCK1 in alcoholic fatty liver disease17,39,40. Therefore, in this study, we found that PCK1 markedly decreased in MAFLD, and PA inhibited PCK1 transcription by upregulating ATF3.
Numerous studies using PCK1 agonists or whole-body Pck1 knockdown mice have verified that PCK1 can affect lipid metabolism41,42. In the present study, liver-specific Pck1 knockout induced significant hepatic steatosis even under normal feeding conditions. This is very important because it is uncommon for single-gene ablation to cause spontaneous steatosis unless a high-fat diet is used. Moreover, mice with liver Pck1 deficiency present aggravated inflammation when fed a high-fat high-fructose diet, contrasting with the results of a previous study showing that whole-body Pck1 knockdown prevents hepatic inflammation43. This discrepancy may be related to differences between diets and animal models, as whole-body Pck1 knockdown may have unexpected effects on glucolipid metabolism. The macronutrient composition of the diet, the duration of feeding, and the genetic background of the animals are all important variables in determining disease severity in preclinical MAFLD models44. To establish an ideal MAFLD model, several attempts have been made to use HFD in combination with fructose and sucrose-enriched drinking water45,46. In this study, we have constructed the MAFLD model using the mouse strain derived from the C57BL/6 J and 129S6/SvEvTac, which may serve as a suboptimal model to investigate advanced stages of MAFLD. In light of the fact that MAFLD prevalence is higher in men than in women at all ages47, we initially used male mice, but not female mice, in the present studies; therefore, the generalizability of the findings to female mice is not guaranteed.
Lipid accumulation is characteristic of steatosis. Emerging evidence indicates that increased fatty acid uptake is associated with lipid accumulation48,49. Previous studies demonstrated that the loss of PCK1 in the liver disturbed hepatic cataplerosis and led to the accumulation of TCA cycle intermediates. The slowed TCA cycle impaired fatty acid oxidation, resulting in fat accumulation in the liver50. Additionally, elevated plasma TGs and FFAs could contribute to body weight gain in L-KO mice. Notably, HFCD-HF/G-induced glucose intolerance and insulin resistance in L-KO mice play essential roles in obesity and whole-body metabolism. In this study, genes involved in fatty acid uptake such as Cd36 and Slc27a1 were highly expressed in L-KO mice. In addition, a lipid droplet-associated protein Cidec was increased by both the chow and HFCD-HF/G diet, and was shown to be upregulated in patients with NAFLD and L-KO mice, suggesting that PCK1 ablation promotes lipid droplet formation51,52. Abnormal levels of metabolites also contribute to TG accumulation in the liver, with the G3P pathway contributing to over 90% of TG synthesis53. As our metabolomics data showed that G3P and PA were significantly upregulated in L-KO mice, PCK1 deficiency may promote hepatic lipid accumulation by enhancing the expression of Cd36, Slc27a1, and Cidec and the levels of metabolic substrates such as G3P and PA. However, the precise mechanism by which PCK1 regulates the G3P pathway and expression levels of Cd36 and Slc27a1 must be further analyzed.
Fibrosis is another characteristic of MAFLD and drives the transition from simple steatosis to NASH. Activation of HSCs through the secretion of profibrotic cytokines, such as TGF-β and PDGF, is a key event in liver fibrosis54. A recent study identified high mobility group protein B1, secreted by fructose-1,6-bisphosphatase 1-deficient hepatocytes, as the main mediator activating HSCs, revealing important crosstalk between hepatocytes and HSCs via paracrine signaling. Herein, PDGF-AA was secreted by PCK1-deficient hepatocytes and acted in a paracrine manner to activate HSCs. Increased deposition of ECM and activation of HSCs were observed in PDGFA-transgenic mice; however, the mechanism mediating PDGF-AA upregulation in fibrosis remains unclear22. Here, we demonstrated that PCK1 deficiency promoted PDGF-AA secretion by activating the RhoA/PI3K/AKT pathway. Mechanistically, PCK1 deletion may increase intracellular GTP levels, thus promoting the activation of RhoA and further activating the PI3K/AKT pathway.
Most Rho GTPases cycle between an active GTP-bound and an inactive GDP-bound form, a process that is regulated by guanine nucleotide exchange factors, GTPase-activating proteins, and guanine nucleotide dissociation inhibitors. Guanine nucleotide exchange factors can activate Rho GTPases by catalyzing the exchange of GDP for GTP when the intracellular concentration of GTP is high55. Several members of the Rho-GTPase family, such as Rac1, RhoA, and RhoC, can be activated by increased concentrations of intracellular GTP56,57,58,59. We found that PCK1 deficiency activated RhoA by increasing the levels of intracellular GTP, therefore activating the downstream PI3K/AKT pathway. Moreover, the genetic and pharmacological disruption of RhoA and AKT1 can effectively mitigate MAFLD phenotypes in L-KO mice. In addition, RhoA and AKT inhibitors can reportedly inhibit the progression of MAFLD through other pathways, such as the NF-κB60, Hippo61,62, and Notch63 signaling pathways. However, off-target effects of these inhibitors cannot be completely ruled out. RhoA and AKT inhibitors are currently only in Phase 3 trials or preclinical studies for the treatment of liver fibrosis or clinical tumors, with therapeutic potential for MAFLD64,65,66.
In conclusion, hepatic PCK1 deficiency promoted lipid deposition and fibrosis in a murine MAFLD model. Moreover, hepatic PCK1 loss activated the RhoA/PI3K/AKT pathway, which increased PDGF-AA secretion and promoted HSC activation in male mice. AKT/RhoA inhibitors reduced progressive liver fibrosis, providing a potential therapeutic strategy for MAFLD treatment.
Methods
Animal models
Animal experiments were approved by the Animal Experimentation Ethics Committees of Chongqing Medical University and performed in accordance with the Guide for the Care and Use of Laboratory Animals. Pck1f/f mice on a 129S6/SvEv background were purchased from the Mutant Mouse Resource & Research Center (MMRRC: 011950-UNC; Bar Harbor, ME, USA) and Alb-Cre mice on a C57BL/6 background were purchased from Model Animal Research Center of Nanjing University (Nanjing, China). To generate liver-specific Pck1-knockout mice (L-KO), Alb-Cre mice were crossed with Pck1f/f mice. Pck1f/f mice from the same breeding step were used as controls (wild-type, WT). Male WT and L-KO mice at 7–9 weeks old were fed the HFCD-HF/G (D12492: 60% Kcal fat, with drinking water containing 23.1 g/L fructose and 18.9 g/L glucose; Research Diets, New Brunswick, NJ, USA) (n = 11 per group) or control chow diet (D12450J: 10% Kcal fat, with tap water; Research Diets) (n = 10 per group) for 24 weeks. Food and drinking water were provided ad libitum. Alb-Cre; Ptenf/f (cPtenf/f) mice (kindly provided by Prof. Yujun Shi, Sichuan University, Chengdu, China) were on a C57BL/6 background and genotyping of the mice was performed as previously described67. L-KO mice were crossed with cPtenf/f mice to generate Alb-Cre; Ptenf/+Pck1f/+ (cPtenf/+Pck1f/+). cPtenf/+Pck1f/+ mice were crossed to breed Alb-Cre; Ptenf/fPck1f/f (cPtenf/fPck1f/f) mice. Littermates that were negative for the Cre transgene were used as WT controls. Primers used for the Pck1, Pten and Cre have been provided in Supplementary Table 2. Male mice were used in all experiments. All mice were housed in temperature-controlled (23 °C) pathogen-free facilities with a 12 h light-dark cycle and humidity (50% ± 10%) conditions.
For AAV8 transduction, AAV8-TBG-control and AAV8-TBG-Pck1 were purchased from the Shanghai Genechem Co., Ltd. (Shanghai, China) and injected via the tail vein following 10 weeks of HFCD-HF/G feeding (2 × 1011 genome copies/mouse). The efficiency of virus infection and expression of PCK1 in mouse hepatocytes were confirmed by western blot analysis. After a total of 24 weeks of HFCD-HF/G feeding, the mice were sacrificed for analysis.
Genetic and pharmacological inhibition of AKT1 or RhoA were performed in vivo. Genetic depletion of mouse RhoA or AKT1 in vivo was conducted using pSECC (#60820; Addgene, Watertown, MA, USA), a lentiviral-based system that combined both the CRISPR system and Cre recombinase. After HFCD-HF/G feeding for 16 weeks, male L-KO mice were injected with pSECC-sgCtrl, pSECC-sgAkt1, or pSECC-sgRhoA through the tail vein at 1 × 109 genome copies per mouse, with two booster injections at 7 day intervals. The pharmacological inhibition of AKT1 or RhoA was conducted using MK2206 or Rhosin. After HFCD-HF/G feeding for 16 weeks, the mice were divided into 3 groups and intraperitoneally injected with vehicle solution (n = 6), MK2206 (AKT inhibitor, 50 mg/kg, every 3 days) (n = 5), or Rhosin (RhoA inhibitor, 20 mg/kg, every 3 days) (n = 6) for 8 weeks. All mice were sacrificed for further study after HFCD-HF/G feeding for 24 weeks.
Liver tissues from patients with NASH
Liver tissue collection was approved by the Institutional Ethics Committees of Chongqing Medical University and Xin Hua Hospital (project license number: XHEC-C-2012-023). Informed consent was obtained from all participants. Paraffin-embedded normal (n = 10) and NASH human liver samples (n = 36) were kindly provided by Dr. Jiangao Fan, Dr. Xiaojun Wang, and Dr. Yalan Wang. Human liver samples with NASH were obtained either by liver biopsy for diagnostic purpose or from surgical liver resections. All liver specimens were evaluated independently by three experienced pathologists, who are blinded to clinical data, according to the NAFLD activity score (NAS), defined as the sum of steatosis, inflammation and hepatocyte ballooning. Patients with a NAS score ≥5 were considered likely to have NASH68. Normal control samples were recruited from samples obtained for the exclusion of liver malignancy during major oncological surgery. Exclusion criteria were the presence of other causes of liver disease, including alcoholic fatty liver disease (>30 g/day for men, >20 g/day for women), chronic infection with hepatitis B and/or C virus, primary biliary cirrhosis, haemochromatosis, autoimmune hepatitis, and Wilson’s disease, as well as the use of anti-obesity, glucose-lowering, and/or lipid-lowering pharmacological treatments. The general characteristics of the NASH human liver samples are listed in Supplementary Table 1.
Cell culture and treatment
All cell lines were grown in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum, 100 µg/mL of streptomycin, and 100 U/mL of penicillin at 37 °C in 5% CO2. All cells were negative for mycoplasma. Short tandem repeat tests were performed to ensure the authenticity of the cells. Bovine serum albumin (BSA, 10%), different concentrations of palmitic acid (PA), 10 µM MK2206 (AKT inhibitor), 40 µM Rhosin (RhoA inhibitor), or blocking antibody against PDGF-AA (2 µg/mL) was added to the medium. For in vitro co-culture assays, the human hepatic stellate cell (HSC) line LX-2 and human hepatocyte line MIHA cells (provided by Dr Ben C.B. Ko, The Hong Kong Polytechnic University, Hongkong, China) were used. LX-2 was pre-cultured in the lower chamber for 12 hours, and PCK1-OE or PCK1-KO cells (with or without indicated treatment) were seeded in Transwell inserts (#3401, Corning, NY, USA) that were subsequently loaded into the LX2-containing wells. The cells and supernatants were harvested for further analysis after 48 h of co-culture.
Isolation of primary mouse hepatocytes
Primary hepatocytes were isolated and cultured from the livers of WT and L-KO mice fed the HFCD-HF/G as described previously69. Briefly, following anesthesia, the inferior vena cava was cannulated and the liver was perfused in situ with 40 mL pre-warmed EGTA solution and 40 mL solution containing 0.35 mg/mL pronase (no. P5147, Sigma-Aldrich, St. Louis, MO, USA), followed by 40 mL solution containing 0.55 mg/mL collagenase (no. V900893, Sigma-Aldrich). After perfusion, the liver was crushed, and hepatocytes were released into the DMEM. The cell suspension was filtered through a 100 μm cell strainer and centrifuged at 50 ×g at 25 °C for 3 min. After washing three times, the cells were suspended in DMEM supplemented with 10 mM glucose, 10% fetal bovine serum, 100 nM insulin (P3376, Beyotime Biotechnology, Shanghai, China), and 100 nM dexamethasone (D8040, Solarbio Life Sciences, Beijing, China), and then plated on 60 mm diameter plastic plates69. After cell attachment, the medium was replaced with serum-free media, and the cells were used for experiments on the following day.
Isolation of primary mouse hepatocytes and primary HSCs
HSCs were isolated from the mice as described previously70. Briefly, after perfusion with solutions containing protease and collagenase, the crushed liver was digested with a solution containing 1% DNase (10104159001, Roche Diagnostics GmbH, Mannheim, Germany), 0.5 mg/mL protease, and 0.55 mg/mL collagenase for 25 min. The cell suspension was filtered through a 70 µm cell strainer, centrifuged at 580 × g for 10 min at 4 °C, and washed twice with Gey’s balanced salt solution (GBSS). The cells were subjected to gradient centrifugation on a 9.7% Nycodenz (1002424, Axis-Shield, Oslo, Norway) to isolate HSCs, which were then plated onto collagen-coated plates. The cells were cultured in DMEM containing 10% (vol/vol) fetal bovine serum and 1% penicillin-streptomycin71.
Construction of adenovirus, lentivirus, and stable cell lines
AdGFP and AdPCK1 adenoviruses were generated using the AdEasy system as described previously5. The PCK1 knockout (PCK1-KO) MIHA cell line was constructed using the CRISPR-Cas9 system (from Prof. Ding Xue, the School of Life Sciences, Tsinghua University, Beijing, China), as described previously5. To knock down ATF3 expression in MIHA cells, three pairs of oligonucleotides encoding short hairpin RNAs (shRNAs) targeting ATF3 or negative control shRNA were cloned into the pLL3.7 vector (from Prof. Bing Sun, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, China). The lentiviruses were obtained by transient transfection of the psPAX2 packaging plasmid and pMD2.G envelope plasmid in HEK293T cells by using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Transfection efficiency was validated by western blotting. Information on the reagents is listed in Supplementary Table 2. The pSECC lentiviral vector cloning and packaging strategy have been described previously27.
RNA extraction and real-time PCR
Total RNA was extracted from the liver tissues or cell lines using Trizol reagent (15596018, Invitrogen, Carlsbad CA, USA) according to the manufacturer’s instructions. RNA was reverse-transcribed using a PrimeScript RT reagent Kit with gDNA Eraser (Cat: RR047A, TAKARA, Shiga, Japan), and qPCR was performed on a Bio-Rad CFX96 machine (Hercules, CA, USA). The mRNA levels of selected genes were calculated after normalization to β-actin by using the 2(-∆∆C(T)) method. Primer sequences are provided in Supplementary Table 3.
Immunoblotting
Mouse liver tissues and cells were homogenized and lysed in Protein Extraction Reagent (P0013, Beyotime Biotechnology) supplemented with protease inhibitor cocktail (04693159001, Roche, Basel, Switzerland). Equal amounts of protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The antibodies are listed in Supplementary Table 4. β-ACTIN protein was used as a loading control.
BODIPY staining
The cells were fixed in 4% formaldehyde for 30 min at room temperature, permeabilized with 0.1% Triton X-100 for 5 min at room temperature, and stained with BODYPY (D3823, Invitrogen) for 60 min. Stained sections were analyzed using a Leica confocal microscope (Leica TCS SP8, Wetzlar, Germany).
CCK8 assay
The cells were seeded at 5 × 103 cells /well in 96-well plates and treated with different concentrations of PA for 24 h. 10 μL CCK-8 solution (#C0005, Topscience, Shanghai, China) was added to each well and incubated at 37 °C for 1 h. The absorbance was measured at 450 nm.
ELISA and G-LISA
Serum insulin (P3376, Beyotime Biotechnology), TNFα (PT512, Beyotime Biotechnology), IL-6 (PI326, Beyotime Biotechnology), and PDGF-AA (SEA523Mu, Cloud-Clone Corp., Wuhan, China) concentrations in the liver tissue and plasma were quantified using mouse ELISA kits according to the manufacturer’s instructions. The levels of secreted PDGF-AA in the cell culture supernatants were determined using a human ELISA kit (SEA523Hu, Cloud-Clone Corp). Active RhoA was detected in a colorimetric RhoA activation assay (G-LISA) (BK124, Cytoskeleton, Denver, CO, USA).
Immunofluorescence
LX-2 cells were fixed in 4% formaldehyde for 25 min and then incubated in 10% normal goat serum for 1 h. The cells were incubated with primary α-SMA antibodies. Specific signals were visualized using secondary antibodies (ZF-0316, Zsbio, Beijing, China). For nuclear staining, the cells were treated with 1 μg/mL DAPI (10236276001, Roche Diagnostics). The samples were detected with a laser-scanning confocal microscope (Leica TCS SP8).
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) and ChIP-quantitative real-time PCR (ChIP-qPCR) assays for MIHA cells were performed as described previously72. Briefly, sonicated chromatin was used for the immunoprecipitation assay. The pre-cleared supernatants were incubated with a monoclonal antibody against ATF3 overnight at 4 °C, followed by a 4 h of incubation with protein A/G agarose beads (LOT: 3460992, Millipore). The purified DNA fragments bound by ATF3 were analyzed using qPCR. IgG and histone H3 were used as negative and positive controls, respectively. The primers used for real-time PCR analysis of PCK1 are listed in Supplementary Table 3.
Glucose and insulin tolerance tests
The glucose tolerance test (GTT) and insulin tolerance test (ITT) were performed at 2 or 1 weeks prior to sacrifice. The animals were fasted for 16 or 6 h, and then glucose solution (2 g/kg body weight) or insulin (0.75 U/kg body weight) was administered via an intraperitoneal injection, respectively. Venous tail blood samples were collected at 0, 30, 60, 90, and 120 min post-administration to assess blood glucose levels using a glucose meter (HGM-114, OMRON, Kyoto, Japan).
Luciferase reporter assay
MIHA cells were treated with 0.2 mM palmitic acid (PA) in 12-well plates for 24 h and then co-transfected with 0.5 μg of pReceiver-M02-ATF3 or shControl or shATF3 plasmid, 0.5 μg of luciferase reporter plasmids pGL3-basic or pGL3-PCK1, and 25 ng of pRL-TK-Renilla (as transfection control) for 48 h. Then, cells were assayed for luciferase activity via the Dual Luciferase Assay Kit (Promega, Madison, WI, USA). All experiments were performed thrice and presented as mean ± standard deviation (SD).
Biochemical analysis
The serum levels of aspartate transaminase (AST), alanine transaminase (ALT), total triglyceride (TG), total cholesterol (TC), and free fatty acids (FFA) were determined with an automated biochemical analyzer (Hitachi 7600, Tokyo, Japan). TG, TC, and FFA levels in the mouse liver tissues were measured with commercial kits according to the manufacturer’s protocol (TG: cat. no. BC0625; TC: cat. no. BC1985; FFA: cat. no. BC0595, Solarbio Life Sciences).
Histological analysis
Paraffin blocks were sectioned into 4 μm slices and used for hematoxylin and eosin (HE) staining, Sirius red staining, and immunohistochemistry (IHC) assay according to standard protocols. Frozen liver tissue sections were stained with Oil Red O (G1260, Solarbio). For pathological grading, all liver specimens were scored by two experienced pathologists according to the NAFLD activity score (NAS), defined as the sum of steatosis (0–3), inflammation (0–3), and hepatocyte ballooning (0–2). An NAS score ≥5 was considered to indicate NASH. Samples were scanned using a slide scanner (Pannoramic DESK, 3D Histech kft, Hungary). For quantitative analysis, the areas of lipid droplets and Sirius red staining were quantified using ImageJ software (version 1.6.0; NIH, Bethesda, MD, USA). Immunohistochemical staining was semi-quantitatively analyzed using the immunoreactive scoring system73. The percentage of positive cells was graded on a scale of 0−4: (0: negative, 1: 0–25%, 2: 26–50%, 3: 51–75%, 4: 76–100%). The signal intensity was scored on a scale of 0–3: 0 = negative; 1 = weak; 2 = moderate; and 3 = strong. Thus, the final immunoreactive score = (score of staining intensity) × (score of percentage of positive cells).
Transcriptomic analyses
Using Trizol reagent, total RNA was isolated from the liver tissues of WT and L-KO mice fed a chow diet or HFCD-HF/G. The RNA quality was checked with a Bioanalyzer 2200 (Agilent Technologies, Santa Clara, CA, USA). cDNA libraries were prepared using an NEBNext® Ultra™ Directional RNA Library Prep Kit, NEBNext® Poly (A) mRNA Magnetic Isolation Module, NEBNext® Multiplex Oligos according to the manufacturer’s instructions (New England Biolabs, Ipswich, MA, USA). Genes with fold-change >2.0 or <0.5 and false discovery rate <0.05 were considered to be significantly differentially expressed. The volcano, heat, and bubble maps were generated using the ‘ggplot2’ or ‘ggpubr’ packages in R (version 3.6.3; The R Project for Statistical Computing, Vienna, Austria). Gene set enrichment analysis was performed using ‘enrichplot’ packages74. The RNA-seq data files have been deposited to the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE162211.
Untargeted metabolomics
Following sacrifice, the livers were snap-frozen in liquid nitrogen and stored at −80 °C until analysis. Untargeted metabolomics was performed using an ultra-high performance liquid chromatography apparatus (Agilent 1290 Infinity LC, Agilent Technologies) coupled to a quadrupole time-of-flight (TripleTOF 6600, AB SCIEX, Framingham, MA, USA) at Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China). Metabolites with a variable importance in projection value >1 were evaluated using Student’s t-test. p < 0.05 was considered to indicate statistically significant results.
HPLC anaIysis of celluIar nucIeotides
Cellular nucleotides were extracted according to published procedures75. Briefly, 1 × 106 cells were washed with phosphate-buffered saline and quenched with liquid nitrogen, and then vigorously mixed with methanol and acetonitrile (1:1, v:v). After incubation on ice for 15 min, the samples were centrifuged at 12,000× g at 4 °C for 10 min. Cellular nucleotides were separated and quantified using a C18 column (Agilent Eclipse XDB-C18, 4.6 × 250 mm, average particle size 5 μm) assembled on the Waters Alliance e2695 Separations Module (Milford, MA, USA). Acetonitrile (5%) and 50 mM KH2PO4 (pH 6.5) containing 10 mM tetrabutylammonium bromide were used as mobile phase A, and acetonitrile was used as mobile phase B. All samples were separated in the mobile phase at a flow rate of 1 mL/min for 30 min at 22 °C. No degradation of individual nucleotides or changes in the ratios of nucleotide mixtures was detected during the experiments. The GTP concentration in the samples was calculated based on the slope of the calibration curves generated using pooled authentic samples (to mimic the matrix), and guanosine-5′-triphosphoric acid disodium salt was used as a standard.
GEO database mining
Raw data in the GSE126848, GSE89632, and GSE135251 datasets were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). R package DESeq2 was used to analyze gene expression levels between different samples, such as NASH vs. health, NAFLD vs. health, and obesity vs. health. Genes showing |log2 fold-change | >1 and false discovery rate <0.05 were considered to present differential expression.
Statistics
Statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad, Inc., La Jolla, CA, USA). Data were represented as the mean ± standard error of the mean unless otherwise stated. Significant differences between the means of two groups were determined using Student’s t-test. One-way analysis of variance was used to determine statistical significance for experiments with more than two groups followed by Tukey’s post hoc test. For correlation analysis, Spearman’s correlation coefficient was used. p < 0.05 was considered to indicate statistically significant results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNAseq data produced in this study were deposited to the public database [Accession GSE162211]. The previously published data sets re-analysed in this study were obtained from [Gene Expression Omnibus (GEO)], through the accession code [Accession GSE126848], [Accession GSE89632], and [Accession GSE135251]16,76,77. The untargeted metabolomic data reported in this article have been deposited in the OMIX, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/omix: accession no. OMIX003062). All other data generated or analyzed in this study are available within the article and its supplementary information files. Source data are provided with this paper.
References
Eslam, M. et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 73, 202–209 (2020).
Eslam, M., Sanyal, A. J. & George, J. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 158, 1999–2014.e1991 (2020).
Schuster, S., Cabrera, D., Arrese, M. & Feldstein, A. E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 15, 349–364 (2018).
Arab, J. P., Arrese, M. & Trauner, M. Recent Insights into the Pathogenesis of Nonalcoholic Fatty Liver Disease. Annu. Rev. Pathol. 13, 321–350 (2018).
Tuo, L. et al. PCK1 negatively regulates cell cycle progression and hepatoma cell proliferation via the AMPK/p27(Kip1) axis. J Exp Clin. Cancer Res. 38, 50 (2019).
Xiang, J. et al. Gluconeogenic enzyme PCK1 deficiency promotes CHK2 O-GlcNAcylation and hepatocellular carcinoma growth upon glucose deprivation. J. Clin. Invest. 131, e144703 (2021).
Xu, D. et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 580, 530–535 (2020).
Santra, S. et al. Cytosolic phosphoenolpyruvate carboxykinase deficiency presenting with acute liver failure following gastroenteritis. Mol. Genet. Metab. 118, 21–27 (2016).
She, P. et al. Phosphoenolpyruvate carboxykinase is necessary for the integration of hepatic energy metabolism. Mol. Cell Biol. 20, 6508–6517 (2000).
Millward, C. A. et al. Phosphoenolpyruvate carboxykinase (Pck1) helps regulate the triglyceride/fatty acid cycle and development of insulin resistance in mice. J. Lipid Res. 51, 1452–1463 (2010).
Hoxhaj, G. & Manning, B. D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88 (2020).
Porstmann, T. et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 24, 6465–6481 (2005).
Huang, X., Liu, G., Guo, J. & Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 14, 1483–1496 (2018).
Chen, J. et al. HIF-2α upregulation mediated by hypoxia promotes NAFLD-HCC progression by activating lipid synthesis via the PI3K-AKT-mTOR pathway. Aging (Albany NY) 11, 10839–10860 (2019).
Chi, Y., Gong, Z., Xin, H., Wang, Z. & Liu, Z. Long noncoding RNA lncARSR promotes nonalcoholic fatty liver disease and hepatocellular carcinoma by promoting YAP1 and activating the IRS2/AKT pathway. J. Transl. Med. 18, 126 (2020).
Suppli, M. P. et al. Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am. J. Physiol. Gastrointest. Liver Physiol. 316, G462–g472 (2019).
Fang, J. et al. Hepatic IRF2BP2 Mitigates Nonalcoholic Fatty Liver Disease by Directly Repressing the Transcription of ATF3. Hepatology 71, 1592–1608 (2020).
Allen-Jennings, A. E., Hartman, M. G., Kociba, G. J. & Hai, T. The roles of ATF3 in liver dysfunction and the regulation of phosphoenolpyruvate carboxykinase gene expression. J. Biol. Chem. 277, 20020–20025 (2002).
Asgharpour, A. et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J. Hepatol. 65, 579–588 (2016).
Liu, X. J. et al. Characterization of a murine nonalcoholic steatohepatitis model induced by high fat high calorie diet plus fructose and glucose in drinking water. Lab. Invest. 98, 1184–1199 (2018).
Kucukoglu, O., Sowa, J. P., Mazzolini, G. D., Syn, W. K. & Canbay, A. Hepatokines and adipokines in NASH-related hepatocellular carcinoma. J. Hepatol. 74, 442–457 (2021).
Thieringer, F. et al. Spontaneous hepatic fibrosis in transgenic mice overexpressing PDGF-A. Gene 423, 23–28 (2008).
Higuchi, M., Masuyama, N., Fukui, Y., Suzuki, A. & Gotoh, Y. Akt mediates Rac/Cdc42-regulated cell motility in growth factor-stimulated cells and in invasive PTEN knockout cells. Curr. Biol. 11, 1958–1962 (2001).
Dou, C. et al. P300 Acetyltransferase Mediates Stiffness-Induced Activation of Hepatic Stellate Cells Into Tumor-Promoting Myofibroblasts. Gastroenterology 154, 2209–2221.e2214 (2018).
Calvayrac, O. et al. The RAS-related GTPase RHOB confers resistance to EGFR-tyrosine kinase inhibitors in non-small-cell lung cancer via an AKT-dependent mechanism. EMBO Mol. Med. 9, 238–250 (2017).
Hodge, R. G. & Ridley, A. J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol Cell Biol. 17, 496–510 (2016).
Sánchez-Rivera, F. J. et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014).
Snaebjornsson, M. T., Janaki-Raman, S. & Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 31, 62–76 (2020).
Vily-Petit, J. et al. Intestinal gluconeogenesis prevents obesity-linked liver steatosis and non-alcoholic fatty liver disease. Gut 69, 2193–2202 (2020).
Yoon, J. C. et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 (2001).
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. & Sanyal, A. J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 (2018).
Samuel, V. T. et al. Fasting hyperglycemia is not associated with increased expression of PEPCK or G6Pc in patients with Type 2 Diabetes. Proc. Natl. Acad. Sci. USA 106, 12121–12126 (2009).
Imai, E. et al. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 10, 4712–4719 (1990).
Lundsgaard, A. M. et al. Mechanisms Preserving Insulin Action during High Dietary Fat Intake. Cell Metab. 29, 50–63.e54 (2019).
Guo, X. et al. The Role of Palmitoleic Acid in Regulating Hepatic Gluconeogenesis through SIRT3 in Obese Mice. Nutrients 14, 1482 (2022).
Wang, Z. & Dong, C. Gluconeogenesis in Cancer: Function and Regulation of PEPCK, FBPase, and G6Pase. Trends Cancer 5, 30–45 (2019).
Jiang, W. et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell 43, 33–44 (2011).
Hai, T., Wolfgang, C. D., Marsee, D. K., Allen, A. E. & Sivaprasad, U. ATF3 and stress responses. Gene Expr. 7, 321–335 (1999).
Tsai, W. W. et al. ATF3 mediates inhibitory effects of ethanol on hepatic gluconeogenesis. Proc. Natl. Acad. Sci. USA 112, 2699–2704 (2015).
Tu, C. et al. Cardiolipin Synthase 1 Ameliorates NASH Through Activating Transcription Factor 3 Transcriptional Inactivation. Hepatology 72, 1949–1967 (2020).
Gut, P. et al. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol 9, 97–104 (2013).
Hakimi, P. et al. Phosphoenolpyruvate carboxykinase and the critical role of cataplerosis in the control of hepatic metabolism. Nutr Metab (Lond) 2, 33 (2005).
Satapati, S. et al. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J. Clin. Invest. 125, 4447–4462 (2015).
Jahn, D., Kircher, S., Hermanns, H. M. & Geier, A. Animal models of NAFLD from a hepatologist’s point of view. Biochim. Biophys. Acta. Mol. Basis Dis. 1865, 943–953 (2019).
Zhou, R. et al. Intestinal α1-2-Fucosylation Contributes to Obesity and Steatohepatitis in Mice. Cell Mol. Gastroenterol. Hepatol. 12, 293–320 (2021).
Tsuchida, T. et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol. 69, 385–395 (2018).
Lonardo, A. et al. Sex Differences in Nonalcoholic Fatty Liver Disease: State of the Art and Identification of Research Gaps. Hepatology 70, 1457–1469 (2019).
Miquilena-Colina, M. E. et al. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut 60, 1394–1402 (2011).
Doege, H. et al. Silencing of hepatic fatty acid transporter protein 5 in vivo reverses diet-induced non-alcoholic fatty liver disease and improves hyperglycemia. J. Biol. Chem. 283, 22186–22192 (2008).
Méndez-Lucas, A. et al. PEPCK-M expression in mouse liver potentiates, not replaces, PEPCK-C mediated gluconeogenesis. J. Hepatol. 59, 105–113 (2013).
Langhi, C. & Baldán, Á. CIDEC/FSP27 is regulated by peroxisome proliferator-activated receptor alpha and plays a critical role in fasting- and diet-induced hepatosteatosis. Hepatology 61, 1227–1238 (2015).
Xu, M. J. et al. Fat-Specific Protein 27/CIDEC Promotes Development of Alcoholic Steatohepatitis in Mice and Humans. Gastroenterology 149, 1030–1041.e1036 (2015).
Alves-Bezerra, M. & Cohen, D. E. Triglyceride Metabolism in the Liver. Compr. Physiol. 8, 1–8 (2017).
Tsuchida, T. & Friedman, S. L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 14, 397–411 (2017).
Zhang, B., Zhang, Y., Shacter, E. & Zheng, Y. Mechanism of the guanine nucleotide exchange reaction of Ras GTPase-evidence for a GTP/GDP displacement model. Biochemistry 44, 2566–2576 (2005).
Wawrzyniak, J. A. et al. A purine nucleotide biosynthesis enzyme guanosine monophosphate reductase is a suppressor of melanoma invasion. Cell Rep. 5, 493–507 (2013).
Hallett, M. A., Dagher, P. C. & Atkinson, S. J. Rho GTPases show differential sensitivity to nucleotide triphosphate depletion in a model of ischemic cell injury. Am. J. Physiol. Cell Physiol. 285, C129–C138 (2003).
Mondin, M. et al. Alterations in cytoskeletal protein expression by mycophenolic acid in human mesangial cells requires Rac inactivation. Biochem. Pharmacol. 73, 1491–1498 (2007).
Bianchi-Smiraglia, A. et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools. Oncogene 36, 84–96 (2017).
Han, M. H. et al. Flavonoids Isolated from Flowers of Lonicera japonica Thunb. Inhibit Inflammatory Responses in BV2 Microglial Cells by Suppressing TNF-α and IL-β Through PI3K/Akt/NF-kb Signaling Pathways. Phytother. Res. 30, 1824–1832 (2016).
Zhao, Y. et al. PI3K Positively Regulates YAP and TAZ in Mammary Tumorigenesis Through Multiple Signaling Pathways. Mol. Cancer Res. 16, 1046–1058 (2018).
Jeong, S. H. & Lim, D. S. Insulin receptor substrate 2: a bridge between Hippo and AKT pathways. BMB Rep. 51, 209–210 (2018).
Gutierrez, A. & Look, A. T. NOTCH and PI3K-AKT pathways intertwined. Cancer Cell 12, 411–413 (2007).
Chien, A. J. et al. MK-2206 and Standard Neoadjuvant Chemotherapy Improves Response in Patients With Human Epidermal Growth Factor Receptor 2-Positive and/or Hormone Receptor-Negative Breast Cancers in the I-SPY 2 Trial. J. Clin. Oncol. 38, 1059–1069 (2020).
Schmid, P. et al. Capivasertib Plus Paclitaxel Versus Placebo Plus Paclitaxel As First-Line Therapy for Metastatic Triple-Negative Breast Cancer: The PAKT Trial. J. Clin. Oncol. 38, 423–433 (2020).
Yoon, C. et al. Chemotherapy Resistance in Diffuse-Type Gastric Adenocarcinoma Is Mediated by RhoA Activation in Cancer Stem-Like Cells. Clin. Cancer Res. 22, 971–983 (2016).
Tong, Z. et al. Pancreas-specific Pten deficiency causes partial resistance to diabetes and elevated hepatic AKT signaling. Cell Res. 19, 710–719 (2009).
Brunt, E. M., Kleiner, D. E., Wilson, L. A., Belt, P. & Neuschwander-Tetri, B. A. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology 53, 810–820 (2011).
Hernández-Alvarez, M. I. et al. Deficient Endoplasmic Reticulum-Mitochondrial Phosphatidylserine Transfer Causes Liver Disease. Cell 177, 881–895.e817 (2019).
Mederacke, I., Dapito, D. H., Affò, S., Uchinami, H. & Schwabe, R. F. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 (2015).
Choi, W. M. et al. Glutamate Signaling in Hepatic Stellate Cells Drives Alcoholic Steatosis. Cell Metab. 30, 877–889.e877 (2019).
Yang, F. et al. GSTZ1-1 Deficiency Activates NRF2/IGF1R Axis in HCC via Accumulation of Oncometabolite Succinylacetone. Embo j 38, e101964 (2019).
Yao, F. et al. A targetable LIFR-NF-κB-LCN2 axis controls liver tumorigenesis and vulnerability to ferroptosis. Nat. Commun. 12, 7333 (2021).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Hannan, J. P. et al. HPLC method to resolve, identify and quantify guanine nucleotides bound to recombinant ras GTPase. Anal. Biochem. 631, 114338 (2021).
Arendt, B. M. et al. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology 61, 1565–1578 (2015).
Govaere, O. et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 12, eaba4448 (2020).
Acknowledgements
We would like to thank Dr. T.-C He (University of Chicago, USA) and Prof. Ding Xue (Tsinghua University, China) for providing the pAdEasy and CRISPR/Cas9 system, respectively. We thank Prof. Youde Cao and Yalan Wang (Chongqing Medical University, China) for providing samples and pathological analysis support. We thank Prof. Yongjun Dang, Hui Zhou and Luyi Huang (Chongqing Medical University, China) for providing the strong technical support for measuring intracellular GTP levels. This work was supported by the National Natural Science Foundation of China (grant no. U20A20392, 82073251, 82273238, 82072286, 82272975), the 111 Project (No. D20028), the Natural Science Foundation Project of Chongqing (cstc2018jcyjAX0254, cstc2019jscx-dxwtBX0019), the Science and Technology Research Program of Chongqing Municipal Education Commission (HZ2021006, KJZD-M202000401), the Future Medical Youth Innovation Team of Chongqing Medical University (W0036, W0101), the Kuanren talents program of the second affiliated hospital of Chongqing Medical University, the Scientific Research Innovation Project for Postgraduate in Chongqing (CYB19168, CYS19193), and the Youth talent fund of Sichuan Provincial People’s Hospital in 2020 (No. 2020QN03).
Author information
Authors and Affiliations
Contributions
N.T., A.H., and K.Wang conceived and designed the study. Q.Y., Y.L., G.Z. and L.T. performed most experiments and analyzed the data. H.D., C.C. and K.Wu conducted bioinformatics analysis. X.P. assisted with mice experiments. X.W., J.F., and Q.P. provided human NAFLD/NASH samples. Q.Y., K.Wang, and N.T. wrote the manuscript with all authors providing feedback. The order of the co-first authors was assigned on the basis of their relative contributions to the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Animal experiments were approved by Animal Experimentation Ethics Committees of Chongqing Medical University and were carried out in accordance with the Guide for the Care and Use of Laboratory Animals. Liver tissue collection was approved by the Institutional Ethics Committees of Chongqing Medical University and Xin Hua Hospital. Informed consent was obtained from all participants.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ye, Q., Liu, Y., Zhang, G. et al. Deficiency of gluconeogenic enzyme PCK1 promotes metabolic-associated fatty liver disease through PI3K/AKT/PDGF axis activation in male mice. Nat Commun 14, 1402 (2023). https://doi.org/10.1038/s41467-023-37142-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-37142-3
This article is cited by
-
Characterization of diseased primary human hepatocytes in an all-human cell-based triculture system
Scientific Reports (2024)
-
Fumigaclavine C ameliorates liver steatosis by attenuating hepatic de novo lipogenesis via modulation of the RhoA/ROCK signaling pathway
BMC Complementary Medicine and Therapies (2023)
-
Comprehensive analysis of epigenetic and epitranscriptomic genes’ expression in human NAFLD
Journal of Physiology and Biochemistry (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.