Abstract
Voltage-gated potassium channels are involved in many physiological processes such as nerve impulse transmission, the heartbeat, and muscle contraction. However, for many of them the molecular determinants of the gating mechanism remain elusive. Here, using a combination of theoretical and experimental approaches, we address this problem focusing on the cardiac hERG potassium channel. Network analysis of molecular dynamics trajectories reveals the presence of a kinematic chain of residues that couples the voltage sensor domain to the pore domain and involves the S4/S1 and S1/S5 subunit interfaces. Mutagenesis experiments confirm the role of these residues and interfaces in the activation and inactivation mechanisms. Our findings demonstrate the presence of an electromechanical transduction path crucial for the non-domain-swapped hERG channel gating that resembles the noncanonical path identified in domain-swapped K+ channels.
Similar content being viewed by others
Introduction
Voltage-gated K+ channels (KV) are membrane proteins that play key roles in many physiological processes, such as generating the nerve impulse, regulating neuronal excitability, shaping the pacemaker in the heart, and controlling muscular contractility1. They are organized as homotetramers of subunits that have six transmembrane segments (S1-S6). The Voltage Sensor Domain (VSD) is formed by segments S1 to S4; the Pore Domain (PD) is composed of S5, P-Loop (a membrane-reentrant region in the protein), Selectivity Filter (SF) that allows only K+ ions to flow through the channel, and S6. Currently, there are two tetramerized architectures identified amongst KV channels: domain-swapped channels (KV1.22,3 and KV7.14) and non-domain-swapped channels (EAG15, hERG6, HCN7, BK8, KVAP9, and Kat110). In domain-swapped channels, the VSD from one subunit contacts the PD from the neighboring subunit whereas in non-domain-swapped channels the VSD and PD from the same subunit are in close contact.
The human ether-à-go-go–related gene11 (hERG or KCNH2) codes for a Kv channel involved in the delayed rectifier current12 (IKr) that determines the plateau period of the cardiac action potential. hERG malfunctions are commonly associated with severe pathologies such as long QT syndrome type 2 (LQTS-2) by loss-of-function mutations (congenital LQTS-2) or channel blockage induced by unspecific interactions with medications (acquired LQTS-2)12,13,14. These conditions have been described to promote arrhythmia and sudden death15.
hERG cycles over three functional states: closed, open, and inactivated. During the peak of cardiac action potential, the channels are mostly in the inactivated state; as the membrane slowly repolarizes, they rapidly recover from inactivation, thus reopening and ceasing the action potential. hERG displays a marked inward rectification, i.e., the currents carried by the channel at depolarizing potentials are relatively small compared with the currents elicited upon repolarization12,16. The mechanism involved in this process is due to an extremely fast pore inactivation (C-type inactivation)17,18,19, which starts before the activation process. These results suggest a possible noncanonical path that directly couples S4 movements to the C-type inactivation20. It has been suggested that this mechanism might depend on a structural constriction of the Selectivity Filter (SF)21 even if its molecular determinants remain elusive.
Recently, an exquisite feature of hERG was unveiled: the channel operates seamlessly without a covalent interaction between the voltage sensor and the pore gate22, strongly suggesting the presence of an alternative path that allows the channel to open excluding the S4-S5 linker loop23,24. Moreover, it has been shown that other regions of hERG can modulate different processes20,25,26,27,28,29,30,31 indicating that its gating mechanism is quite complex and not fully understood.
Ion channels can be viewed as allosteric machines32, transferring information from a sensor (VSD) to an actuator (PD). In this work, to reveal the electromechanical VSD-PD coupling mechanism in the hERG channel, we combined MD simulations to network analysis33,34,35, verifying the existence of a noncanonical route of motion that propagates across the channel. We theoretically identified a chain of residues coupling the VSD to PD that involves S4\S1 and S1\S5 subunit interfaces. Then, we validated these predictions using electrophysiological techniques revealing a path very similar to that involved in the noncanonical gating of domain-swapped channels33,36,37,38. These results provide insights into the activation and inactivation mechanisms of hERG and demonstrate the presence of a noncanonical electromechanical coupling in non-domain-swapped channels.
Results
Electromechanical noncanonical paths predicted by network analysis
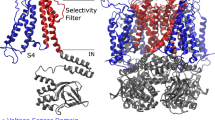
Molecular Dynamics (MD) simulations of the hERG wild type (WT) in the open and closed states were run starting from the experimental structure solved by MacKinnon’s lab6 and the system with gating charge Qg = 8e predicted in Costa et al. 202224, respectively. To characterize the allosteric mechanisms of the VSD-PD coupling, the two systems were represented as networks where nodes coincide with protein residues and edges with the interactions between pairs. Each edge was assigned a weight expressed as \({w}_{{ij}}=-{{\log }}\left({C}_{{ij}}{M}_{{ij}}\right)\), which quantifies the electromechanical coupling in terms of contacts between residues and correlations of their motion, computed from MD runs. Dijkstra’s algorithm39 was used to determine the shortest paths between S4-S6 and S4-SF (for more details see Methods). Two allosteric paths were determined (Fig. 1) where the S4\S1 and S1\S5 subunit interfaces are prevalently connected by hydrophobic interactions (Supplementary Fig. 1). Analyzing the closed state trajectories, besides retrieving the canonical S4 → L45 → S6 path not discussed here but reported in Supplementary Fig. 2, we noted that displacements of the voltage sensor helix S4 propagate to helix S1. Subsequently, they pass on helix S5 and, finally, reach helix S6 in the PD determining the channel opening (blue arrows in Fig. 1). Similarly, in the open state trajectories, motion propagates from helix S4 passing through S1 and S5 following the same route as previously described for the closed trajectories. In this latter case, motion subsequently propagates to the SF via the P-Loop (yellow arrows in Fig. 1). These paths resemble the noncanonical VSD-PD coupling identified in the domain-swapped Shaker K+ channel 36,37,38 and they are expected to be involved in the activation and inactivation mechanisms of hERG, respectively. Interestingly, the presence of the noncanonical activation path that coexists with the classical path involving the S4-S523,24 would explain the opening of the channel even if a disconnection between the VSD and the PD had been introduced22,40.
Panels a and b show the intramembrane and extracellular views of the open state from MD simulations of the wild type24 colored by domains: VSD in blue, PD in red and cytoplasmic domain in gray. Panel c schematizes the noncanonical paths found by simulations and involved in the activation (blue arrows) and in the inactivation (yellow arrows) that couple S4-S6 and S4-SF, respectively. The residues identified using the network analysis with the highest values of betweenness centrality (see also Supplementary Table 1) that are involved in the noncanonical gating paths are indicated by a label and colored by individual amino acids using VMD 1.9.470 (ResName style). The blue and yellow paths overlap until segment S5, where they diverge. To avoid confusion between the canonical and the noncanonical electromechanical coupling involved in the activation, we did not study residues on helix S6 since it would be difficult to distinguish the resulting effects on the process that depend on the canonical mechanism from those depending on the noncanonical mechanism.
Mutation of residues involved in the noncanonical paths
The role of each residue in the predicted noncanonical paths was quantified by performing betweenness centrality41 (BC) calculations on the wild-type systems. Residues with high values of BC (Fig. 1 and Supplementary Table 1) act as hubs in the electromechanical coupling between VSD and PD and are predominantly located on S1, S5, and the P-Loop suggesting a key role of the S1/S5 interface in the hERG noncanonical gating. Based on these theoretical predictions, the individual contribution of key residues was tested using electrophysiological assays. Residues T425, P426, A527, H562, W563, A565, W568, A614, Y616, F617, and T618 were mutated to leucine. We chose a leucine-scanning mutagenesis to perturb the identified chain and assess the role of residues in the activation and inactivation paths because the standard alanine and valine scanning approaches led to insufficient expression of the mutants. More specifically, it has been reported that alanine mutations on residues W568, Y616, F617 and valine mutations on residues A565, W568, and A614 led to channels with lack of expression20,42. Mutations of residues on the S4 helix (L524R, L529H, and L532H) were also produced as they had already been identified to affect the Shaker noncanonical gating mechanism36 (see the sequence alignment in Supplementary Fig. 3 between hERG (UniProt ID: Q12809) and Shaker (UniProt ID: P08510) using Clustal Omega43). Finally, A614 was mutated into glycine because bulkier residues such as valine20 or leucine (A614L—this work) did not exhibit constructs able to generate currents. Due to the lack of expression, we were not able to characterize the following residues when mutated to leucine: P426, H562, A565, and A614.
Voltage dependence of the activation process
First, we set to determine whether the mutations affected the voltage dependence of the activation process. Since hERG exhibits a marked fast rectification/inactivation, the voltage dependence of activation was calculated by the relative amplitude of the peak tail currents (black arrow in Fig. 2a) plotted against the membrane potential (G-V curves). We categorized the effects of mutations on the voltage dependence of activation as left-shift and right-shift mutants (Fig. 2 and Table 1). Left-shift mutants favor the open state of the channel lowering the activation energetic barrier: they were L524R, A527L, L529H, L532H, and W563L. The magnitude of the shift in G-V varied, with the largest left shift happening in L529H and W563L, about −30 mV, whereas the smallest left shift occurred in A527L, about −6 mV (Table 1 and Fig. 2). The mutants that have a right shift favor the closed state of the channel: T425L (~20 mV); A614G (~3 mV) and T618L (~5 mV) show only a minor right shift. The time course of the current exhibited different phenotypes and three stand out. The mutants A527L (S4) (Fig. 2d), W563L (S5) (Fig. 2g) and T618L (P-Loop) (Fig. 2i) exhibited a marked peak (red arrows) with a large outward current compared to their inward tail and with the wild type channels. These results suggest that the inactivation/rectification of these channels were relieved, even though the voltage dependence of these channels were different.
a–i are the representative current traces for the WT, T425L, L524R, A527L, L529H, L532H, W563L, A614G, and T618L, respectively. Inset in a is the voltage protocol used and the black arrow denotes the time where the tail currents were measured at –100 mV to obtain the G-V curves. j Voltage dependence of activation (G-V curves) for the channels studied. The experimental data were fitted using Eq. 6 and the best fitted values were displayed in Table 1. The holding potential was –100 mV. Data are presented as mean ± SD (WT (n = 4), T425L (n = 4), L524R (n = 6), A527L (n = 4), L529H (n = 4), L532H (n = 4), W563L (n = 4), A614G (n = 4), and T618L (n = 4), where n is the number of biologically independent cells).
Voltage dependence of the inactivation process
Three of the studied mutants exhibited no inactivation: W568L, Y616L, and F617L (Fig. 3). Residue W568 was previously identified to couple/interact with the selectivity of the P-loop20; here our network analysis showed that it is also involved in the noncanonical path, accounting for its lack of inactivation (Fig. 3a) as previously shown for W568L20. Similarly, mutants Y616L and F617L exhibit no inactivation, and a marked right shift in the G-V curves (Fig. 3b–d). These results suggest that these residues are involved in both hERG activation and inactivation.
a–c are the representative current traces for the W568L, Y616L, F617L, respectively. Inset in a is the voltage protocol used. For mutant Y616L the highest voltage applied was +120 mV. Because those mutants are right shifted, we had to modify the voltage protocol used in Fig. 2a to account for the shifts. d The peak current normalized by its maximum plotted against the membrane voltage. Data are presented as mean ± SD (WT (n = 3), W568L (n = 4), Y616L (n = 6), F617L (n = 3), where n is the number of biologically independent cells).
Next, since the chain of residues identified via MD simulations connects the VSD to the SF that is involved in the inactivation process21, we hypothesized that the mutants tested here would affect the inactivation process if they were involved in a noncanonical path. As predicted, electrophysiological measurements confirm that they affect the inactivation (Fig. 4 and Table 1). L524R, A527L, L529H, L532H, and A614G showed a left shift with different magnitudes in the voltage-dependence of inactivation (Table 1). T425L showed a 27 mV right shift while W563L and T618L displayed no change in the voltage dependence of the inactivation. Interestingly, the S4 mutant L529H and the S5 mutant W563L exhibited similar behavior: the G-V curve is shifted to the left by ~−30 mV, but the voltage dependence of inactivation is left shifted only by ~−15 mV for L529H or does not have shift as for W563L. The opposite happened to the S4 mutation L532H, where the G-V curve is shifted to the left by ~−10 mV, but the inactivation is shifted to the left ~−25 mV. Another interesting result concerns the S1 mutant T425L for which the G-V is right-shifted by 20 mV, but the inactivation is right shifted by ~27 mV. The shifts in the voltage dependence of activation and inactivation were distinct regarding the amplitude and directionality, suggesting an impairment in the coupling between activation and inactivation. Therefore, they support the evidence for the noncanonical path proposed here. Since mutants W568L, Y616L, and F617L (Fig. 3) did not exhibit signs of inactivation we did not further explore the inactivation effects of these mutants.
a Currents elicited using a classical three-pulse voltage protocol shown inset in a. A voltage pulse to +20 mV (P1) was applied for 1 s to fully activate/inactivate the channel, followed by a voltage pulse (P2) applied to different potentials from −180 to +60 mV for 30 ms. This pulse was used to remove inactivation without allowing sufficient time for significant deactivation to occur. A final pulse (P3) was then applied to +20 mV for the reopening and allowed the channels to reenter into the inactivated state. For better appreciation of the currents elicited by pulses 2 and 3, we expanded the time window of the current traces (dashed grey square in a). b–j are, respectively, the representative expanded time window for WT, T425L, L524R, A527L, L529H, L532H, W563L, A614G, and T618L current traces. j Voltage dependence curve for inactivation. The voltage dependence of inactivation was assessed by the peak currents elicited by P3 (red arrow in b and normalized by its maximum (I/IMAX). For hyperpolarized potentials, <−120 mV, the channels start to deactivate which reduces the current elicited by P3, which underestimates the peak current. In order to account for this process, we corrected by extrapolating the currents back to the start of P2 from the falling phase of the currents elicited by P218. For the right-shifted mutant T425L, for the mutant A527L P1 and P3 were set to +80 mV to account for the shift in the G-V curve and to fully inactivate the channel. Similarly, for T618L mutant, P1 and P3 were set to +100 mV to fully inactivate the channel. k Voltage dependence data for mutants. The voltage dependence data were fitted using Eq. 7 and the best-fitted values were displayed in Table 1. Data are presented as mean ± SD (WT (n = 4), T425L (n = 6), L524R (n = 4), A527L (n = 5), L529H (n = 3), L532H (n = 5), W563L (n = 3), A614G (n = 5), and T618L (n = 4), where n is the number of biologically independent cells).
Comparison between theoretical predictions and experiments
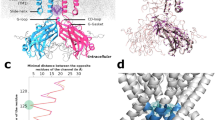
The theoretical predictions on the importance of residues involved in the noncanonical paths were verified by experiments as confirmed by plotting the betweenness centrality of each mutated residue along the WT noncanonical paths against the free-energy perturbation of activation (ΔΔGG) and of inactivation (ΔΔGI) (see Methods). From the plots shown in Fig. 5, it is possible to appreciate correlations between BC of the wild-type residues with |ΔΔGG| for activation and |ΔΔGI| for inactivation (Fig. 5a, b). To characterize microscopically the individual role of these residues, mutants were also produced by computational mutagenesis. We analyzed 150 ns simulations with the same network-theoretical approach. In general, the noncanonical paths involved in both activation and inactivation were not qualitatively modified. However, there were significant differences in terms of path lengths (Supplementary Table 2), which account for an exponential improvement or impairment of information transfer. Since the mutations did not substantially alter the contact patterns, these differences depend only on how effectively the motion propagates between VSD and PD. To provide a microscopic interpretation of the experimental data, we also plotted the minimal path length variation (Δdmin) of each mutant along the noncanonical paths against ΔΔGG and ΔΔGI which are again correlated. These plots highlight that lower efficiency in the motion propagation (higher Δdmin) correlates with an increasing impairment of the activation/inactivation mechanisms (higher ΔΔG). Similarly, a more efficient transfer of motion (lower Δdmin) correlates with improved gating as measured by experiments (lower ΔΔG). These data demonstrate a fair agreement between computational and experimental results reinforcing the notion that the residues identified are part of the noncanonical electromechanical coupling in hERG.
Panels a and b refer to betweenness centrality vs | ΔΔGG | and betweenness centrality vs | ΔΔGI | , respectively. Panels c and d refer to Δdmin vs ΔΔGG and Δdmin vs ΔΔGI, respectively. Green lines are linear fits to the data computed by the regression analysis derived by n = 8 independent samples (i.e., points in each panel) and “r” is the corresponding correlation coefficient. The values of ΔΔGG and ΔΔGI are presented on Table 3. They were evaluated from Eqs. 8 and 9 from the fitted values obtained from Figs. 2j and 4k, respectively. The procedure to obtain ΔΔGG and ΔΔGI is described in Methods. Data are presented as mean values ± standard error (SE) associated to the energies and mean ± SD for betweenness centrality and the average minimal path length variation. The number of cells of biologically independent experiments associated with calculations of the activation and inactivation energies and the standard error associated to them are shown in Figs. 2j and 4k, respectively.
Figure 5c shows a strong correlation between mechanical coupling and effectiveness of activation, as shown by the correlation coefficient r = 0.905. For inactivation, Fig. 5d shows a moderate correlation that improves if the main outlier A614G is excluded by the regression analysis. The correlation of computational and experimental data is somewhat reduced for inactivation possibly because of the smaller values of zIFVI1/2 associated to smaller variations of the gating charge zI and larger relative errors on ΔΔGI. Since glycine is known to generate bends in the helix, we observe that the glycine replaced at A614 determines a marked bending of the P-Loop perturbing the structural arrangements and inducing a constriction of the SF (Supplementary Fig. 4). Hence, inactivation is favored in this mutant when compared to the wild type (Fig. 4j, Tables 2 and 3) even if this is not apparent in simulations, because it does not lie on the main path. Finally, simulations predict that mutants W568L, F616L, and Y617L (Supplementary Table 2) do not exhibit inactivation consistently with experimental data (Fig. 3), demonstrating the robustness of the current approach.
Discussion
Several recent works have highlighted the presence of a new mechanism that couples the VSD to the PD excluding the S4–S5 linker in the domain-swapped Shaker channels36,37,38. This alternative path was defined as “noncanonical” because it extends beyond the classical electromechanical coupling based on the action of the S4–S5 linker. Recently, computational studies have shown the presence of a similar alternative path in the hERG channel, suggesting that a noncanonical activation mechanism can also play a role in non-domain-swapped channels24. Here, we combined theoretical and experimental approaches to characterize this noncanonical electromechanical coupling mechanism in the hERG channel.
First, using MD simulations combined with a network analysis, we identified two paths, not related to the S4-S5 linker, that couple the VSD-PD (activation) and VSD-SF (inactivation), where the S4\S1 and S1\S5 subunit interfaces are the connecting points between VSD and PD. Interestingly, the S4/S5 interface was shown to play a role also in domain-swapped K+ channel36,37,38. For the activation path, simulations of the closed system with Qg = 8e produced in Costa et al. 202224 were analyzed and a noncanonical path that follows the S4 → S1 → S5 → S6 route was found. The choice to predict the activation path on the closed state of hERG relies on a large body of experimental evidence suggesting that the open state is intrinsically more stable than the closed state44,45,46,47. It has been shown that the VSD must exert work to close the pore44,48,49,50 and, when no electric field is applied, the channel “falls” into the open state51. These results suggest that the noncanonical communication paths for activation are not present in the open state, and they gradually build as the channel approaches the closed state. Determining the weights of the protein network from simulations of the closed state, therefore, seems the best strategy to capture the features of a fully developed communication network. Despite the uncertainty on the structure of the closed state, which was generated via homology modelling and steered MD simulations, the high correlation in Fig. 5c suggests that the noncanonical communication path for the activation revealed in this work may actually be the underlying mechanism that explains the seamless function of the channel without a covalent interaction between the voltage sensor and the pore gate22,40. It is important to stress that the representation of the hERG channel employed in our work may be a simplified model of the system. For instance, while we consider a single closed state, in Markov State models, hERG requires a representation with three closed states, an open state, and an inactivated state52. Within this framework hERG gating could be much more complex, possibly including different pathways for the transition between the different closed states and the open state. Even if this scenario is fascinating, the unavailability of experimental structures for the three closed states currently precludes computational characterization using our MD/network approach. The consistency between computational predictions and experiments, however, suggests that our model, though possibly simplified, captures the essential features of the hERG gating mechanism.
The network weights for the computation of the inactivation paths were derived from equilibrium simulations of the open state because the structure of hERG inactivated state is currently rather elusive. Even though a recent computational work21 has suggested a mechanism of constriction of the selectivity filter, more experimental data are still necessary to fully elucidate this mechanism. Moreover, it seems that the structure of the inactivated state may be very similar to that of the open state, consistently with the extremely fast open-to-inactivated transition6. Given the current lack of structural information, we decided that using the open state to predict the inactivation path was the safest approach. In this case, we identified a kinematic chain of residues that electromechanically propagates the movements of VSD and the constriction of the SF following the S4 → S1 → S5 → P-Loop→SF route relevant for the C-type inactivation.
The importance of residues in the electromechanical couplings was quantified by computing the betweenness centrality of each residue. As will be shown in the following, this a priori analysis of the wild type is a convenient and reliable way of guiding experimental and computational mutagenesis. We showed in Fig. 1 that residues P426 and T425 on helix S1 were identified to play a key role in the noncanonical activation and inactivation paths. These results agree with the work by Phan et al.29 where it has been shown that P426A and T425A perturbed the voltage dependence and the kinetics of the activation and the recovery from the inactivation. This evidence suggests that mutations of S1 residues break the connections between the VSD and the PD confirming the crucial role of helix S1 in coupling the VSD to the PD and completing the view proposed by Wang et al.28 of how the motions of helices S4 and S5 are connected. Similarly, helix S5 mediates the activation and the inactivation connecting helix S1 to helix S6 via W568, A565, W563, and H562. These results confirm the role of helix S5 on the hERG gating, as already shown in previous works20,42,53.
To confirm the theoretical predictions on the wild type, mutants were computationally and experimentally produced using leucine-scanning mutagenesis or structurally related mutations that have already been shown to be involved in the noncanonical path in Shaker channels36. In the corresponding simulations of the mutants, the communication paths were surprisingly similar to those of the wild-type channel, but with significantly different efficiencies (path lengths) in connecting the VSD to the PD. For activation, W563L, L529H, A527L, and L524R had a path length smaller than the wild type suggesting that the mutations increase exponentially the efficiency of the VSD-PD coupling path. Conversely, A614G, T618L, and T425L showed a weaker electromechanical coupling because they display longer path lengths. For inactivation, in L524R and A527L there were more efficient paths while in W563L, T618L, L529H, L532H, and T425L the inactivation path was less efficient. To provide unequivocal evidence of the noncanonical path we measured from experimental curves the free-energy perturbation of each channel for the activation and inactivation and related them with the change in the path length in the corresponding mutants. Our analysis was based on the correlation between the betweenness centrality vs |ΔΔGI| and |ΔΔGG|, and Δdmin vs ΔΔGI and ΔΔGG, rather than establishing a qualitative threshold for the ΔΔGI or ΔΔGG, for instance <1 kcal/mol. The reason for that is that the inactivation process has less voltage sensitivity (zI) when compared to the activation curves (zG ~ 3 and zI ~ 1), which provides lower values for ΔΔGI compared to ΔΔGG for the same shift in the voltage dependence (V1/2). Therefore, we reasoned that a correlation as that shown in Fig. 5 would provide a broader assessment of the contribution of each residue in the motion propagation through the noncanonical path and a quantification of the agreement between theoretical and experimental data. In this context, the correlations presented in Fig. 5 show that the experimental and theoretical results are in good agreement. The main exception is represented by the outlier A614G whose behavior is likely to depend on the presence of indirect effects on the structure of the protein backbone that could not be detected by the network analysis.
Interestingly, residues with high betweenness centrality in simulations, a measure of their relative importance in the path, affect prominently the gating mechanism also in experiments. Indeed, the scatter plots in Fig. 5a–d reveal that the greater the betweenness centrality values the higher the absolute values of ΔΔG for activation and inactivation. The mutagenesis study shows that residues in the noncanonical path act as hubs in the communication network affecting the activation and inactivation mechanisms. There is a clear advantage of using MD simulations and network analysis to make predictions on the paths and on the individual role of each residue. Information on the wild-type system based on BC represents a sufficient but not necessary condition for a residue to affect the gating mechanism. Thus, a residue with high BC value most likely will affect that mechanism. The main limitation of this computational tool is that it cannot guarantee that mutations on residues showing low BC values will not have an impact on activation or inactivation due to a plethora of indirect effects (e.g., A614G discussed in the Results). In this context, the correct way to use our computational results is to significantly narrow down the search and guide experimental mutagenesis.
Altogether, our work demonstrates the presence of an alternative allosteric gating mechanism in the hERG channel that involves the S4\S1 and S1\S5 subunit interfaces. The present work shows that this so-called noncanonical path plays a central role in the inactivation thus completing the picture sketched in our previous contribution24 where we showed that in the activation process, the noncanonical path coexists and complements the canonical one. The relative contribution of the two paths to the activation/deactivation mechanism, however, still needs a quantitative characterization through further experiments and simulations. Furthermore, these paths resemble those recently proposed for domain-swapped channels36,37,38, suggesting the presence of a common underlying mechanism of electromechanical coupling across the superfamily of Kv channels.
Our results shed light on the elusive activation and inactivation mechanisms of non-domain-swapped channels which might serve as a basis for studying the mechanisms leading to inherited and induced channelopathies.
Methods
MD simulations
The open and closed states of hERG channels were generated, as detailed in Costa et al. 202224, and simulated for 500 ns in the NPT ensemble. hERG open state was produced from the experimentally solved structure (PDB ID 5VA26), while the closed state was generated through homology modeling from the template EAG1 (PDB ID 5K7L5) and Steered MD simulations. The latter was run to pull helix S4 in the closed position corresponding to a gating charge Qg = 8e, as the experimental value recorded by Zhang et al.54, 6.4e, increased by 20% to account for the underestimation affecting the experimental technique55,56. Mutants were produced from the pre-equilibrated wild-type system. At first, they were equilibrated for 6.5 ns in the NPT ensemble applying a time-varying harmonic restraint on each mutated residue and on its neighbors within a cut-off distance of 5.0 Å. The force constant, initially set to 10 kcal/mol/Å2, was decreased by 2 units every 0.5 ns of this simulation and then the systems were equilibrated for 150 ns in the NPT ensemble. All simulations were run with NAMD 2.1457 using the ff14SB force field for the protein58, the Lipid17 force field for the lipids59 and the TIP3P water model60. In all simulations, pressure was kept at 1.01325 bar by the Nosé-Hoover Langevin piston method61,62 and the temperature was maintained at 303.15 K by a Langevin thermostat with damping coefficient of 1 ps−1. Long-range electrostatic interactions were evaluated with the smooth PME algorithm63 with a grid space of 1 Å. For short-range non-bonded interactions, a cut-off of 12 Å with a switching function at 10.0 Å was used. The integration time step was 2 fs.
Network analysis
The motion propagation at the basis of the VSD-PD and VSD-SF coupling mechanisms was studied representing the protein as a graph64 where nodes correspond to the protein residues and edges to the interactions between pairs. Each edge was assigned a weight expressed as:
here, \({C}_{{ij}}\) is a semi-binary contact map and \({M}_{{ij}}\) is a matrix that quantifies the motion correlation of residues \(i,j\). As shown in Supplementary Fig. 5, \({C}_{{ij}}\) was computed with a truncated Gaussian kernel:
where \({d}_{{ij}}\) is the distance between the Cα of the \(i\) and \(j\) residues and \(c\) is the cut-off distance set to 7.0 Å. The width \(\sigma\) of the gaussian kernel was chosen so as to attain a negligibly small value of the kernel at \({d}_{ij}=10\) Å. Specifically, we imposed \(K({d}_{{cut}})={10}^{-5}\)attaining \(\sigma=1.48\). The contact map is computed by averaging the value of the kernel over all the frames of the trajectory:
The mutual information \({M}_{{ij}}\) of two random variables is a measure of their reciprocal independence or coupling and was used to quantify the motion correlation of two residues. Defining \({d}_{i}\) and \({d}_{j}\) as the displacement of the center of mass of the side chain with respect to its average position, the mutual information is:
and it quantifies the loss of uncertainty on the position of residue \(i\) knowing the position of residue \(j\). We used normalized mutual information \({{M}\prime}_{{ij}}=\frac{{M}_{{ij}}}{{H}_{{ij}}}\) where \({H}_{{ij}}\) represents the Shannon entropy65 of the variables \({d}_{i}\) and \({d}_{j}\). Since both the normalized mutual information and the semi-binary map assume values in the range [0, 1] the weights \({w}_{{ij}}\) will be non-negative, satisfying a requirement of Dijkstra’s algorithm39 that we used to compute the minimal paths between key regions centered on S4 and S6 helices and on the SF. Residues belonging to the key regions are reported in Supplementary Table 3. The dmin values correspond to the lowest values computed from Eq. 1. The betweenness centrality of each residue was computed with Brandes’s algorithm41 as implemented in the NetworkX 3.0 library66. A step-by-step example of network analysis is described in Supplementary Information, section “Step-by-step example of network analysis”.
Statistics and reproducibility
The variability of the network weights \({w}_{{ij}}\) was determined using a block analysis67,68. The simulations were split into \({N}_{B}\) sub-trajectories of 25 ns, long enough to consider the \({w}_{{ij}}\) computed in the different blocks as uncorrelated measurements. The standard deviation for each element of the matrices was computed:
where \({w}_{{ij}}^{B}\) is the value of the weight computed in block \(B\) while \({\bar{w}}_{{ij}}^{B}\) is the average over all blocks.
Site-directed mutagenesis
Human hERG K+ channel cloned into SP64 vector (kindly provided by Dr. Eduardo Perozo—University of Chicago). Mutations were performed using Quick-change II technology (Stratagene, La Jolla, CA), together with custom primers from Integrated DNA Technologies (Integrated DNA Technologies, Inc., Coralville, IA). hERG cDNA and its mutants were sequenced to ensure accurate DNA sequencing, linearized by endonuclease EcoRI (New England Biolabs, Ipswich, MA) and cleaned up with a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Bethlehem, PA). In vitro transcription kits were used to transcribe cDNA and generate cRNA (SP6 RNA expression kit; Ambion Invitrogen, Thermo Fisher Scientific, Waltham, MA).
Oocytes preparation
Oocytes were harvested from Xenopus laevis in accordance with experimental protocols #71475 approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC). The follicular membrane was digested using collagenase type 2 (Worthington Biochemical Corporation, Lakewood, NJ) – 2 mg/ml and supplemented by bovine serum albumin (BSA). Following the follicular membrane digestion, oocytes were incubated in standard oocytes solution (SOS) containing, in mM: 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 0.1 EDTA, 10 HEPES and pH set to 7.4 with NaOH. SOS was supplemented with 50 µg/ml gentamycin to avoid contamination during incubation. After 6 to 24 hours of harvesting, defolliculated oocytes stage V-VI, were injected with cRNA (5 to 100 ng diluted in 50 nl of RNAse free water) and incubated at 18 °C prior to recording. Unless otherwise stated chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Electrophysiological recordings
Ionic currents were recorded using cut-open oocyte voltage-clamp (COVC) method69. Currents were acquired by a setup comprising a Dagan CA-1B amplifier (Dagan, Minneapolis, MN) with a built-in low pass 4-pole Bessel filter. A 16-bit A/D converter (USB-1604, Measurement Computing, Norton, MA) was used by acquisition and controlled by an in-house software (GPatch64MC). Data was sampled at 1 MHz, digitally filtered at Nyquist frequency and decimated for the desired acquisition rate. Capacitive transient currents were compensated using a dedicated circuit. The voltage-measuring pipette was pulled using a horizontal puller (P-87 Model- Sutter Instruments, Novato, CA). For ionic currents measurements, the external solution was composed by (mM): KOH 12, CaOH2, HEPES 10, EDTA 0.1, NMDG (N-Methyl-D-glucamine) 108, pH 7.4 (adjusted with MES—Methanesulfonic acid). The internal solution was composed of (mM): KOH 120, EGTA 2, HEPES 10, pH7.4 (adjusted with MES—Methanesulfonic acid). The holding potential was set to −100 mV. Recordings were performed at room temperature (~ 17–18 °C).
Data analysis
The voltage dependence of the channel conductance was taken by tail currents. After a long depolarizing pulse to different membrane potentials (P1), the membrane was set to a hyperpolarized voltage of −100 mV (P2). The currents elicited by P2 were normalized by its maximum of each experiment, averaged, and plotted against the membrane voltage of pulse P1 in order to obtain a conductance-voltage (G-V) curve. The G-V curves were fitted using a two-state model with the following equation:
where \({z}_{G}\) is the apparent valence of the charge times the fraction of the electric field of the transition and \({V}_{G1/2}\) is the voltage for 50% of the maximal conductance.
The steady-state inactivation currents were assessed using a triple-pulse protocol: a condition depolarized pulse (P1) for 1 s, followed by short (30 ms) hyperpolarizing pulses (P2) varying from –180 mV to +60 mV or +100 mV for some mutants (increments of 10 mV) and another test pulse (P3) for 0.6 s. The peak currents elicited by P3 were corrected by extrapolation of the decay phase of the currents elicited by P218. The relative peak currents (I/IMAX) were plotted against the voltage of pulses P2. The inactivation voltage dependence was assessed by the I/IMAX curves for each mutant using a two-state model with the following equation:
where \({z}_{I}\) is the valence of the apparent charge times the fraction of the electric field of the transition and \({V}_{I1/2}\) is the voltage for 50% of the maximal inactivation.
To evaluate the effect of leucine on the identified noncanonical path, we calculated the perturbation energy from all the single mutants and wild type. Thus, we used the activation free-energy (\({\varDelta G}_{G}\)) for each channel evaluated by the \({V}_{G1/2}\) and \({z}_{G}\) from the G-V curves, using the following equation:
Similarly, we used the same idea to calculate the perturbation energy to the inactivation free-energy (\({\varDelta G}_{I}\)) using the following equation:
To calculate energetic perturbation of each mutant on the free-energy of the activation or inactivation process (\({\varDelta \varDelta G}_{G}\) or \({\varDelta \varDelta G}_{I}\), respectively), we calculated the difference between the free-energy of mutant (\({\varDelta G}_{{{{{\rm{Mutant}}}}}}\)) from the wild type (\({\varDelta G}_{{WT}}\)) channels for activation or inactivation process using the equation:
The standard error (SE) associated to the energies of activation or inactivation \(\delta \triangle \triangle G\) were calculated using the following equations:
Matlab (The MathWorks, Inc., Natick, MA) and Origin 9.0 (Origin Lab Corporation, Northampton, MA) were used for calculating G-Vs, Q-Vs, plotting and fitting the data. Data analysis were also performed using a program written in-house (Analysis).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon request. The MD trajectories that support the findings of this study are available in Zenodo with the identifiers: [https://doi.org/10.5281/zenodo.7100860], [https://doi.org/10.5281/zenodo.7371824] and [https://doi.org/10.5281/zenodo.7372042]. The contact maps are available in Zenodo with the identifier: [https://doi.org/10.5281/zenodo.7470218]. Accession codes of the structures used to produce the open and closed states, respectively: hERG open state PDB ID 5VA2 and EAG1 closed state PDB ID 5K7L. Accession codes used for the sequence alignment: hERG UniProt ID Q12809 and Shaker UniProt ID P08510. The data underling Figs. 2–5 are provided as Source Data File. Source data are provided with this paper.
References
Hille, B. Ion Channels of Excitable Membranes. (Sinauer Associates, 2001).
Long, S. B., Campbell, E. B. & MacKinnon, R. Crystal structure of a mammalian voltage-dependent shaker family k+ channel. Sci. (80-.) 309, 897–903 (2005).
Chen, X., Wang, Q., Ni, F. & Ma, J. Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc. Natl Acad. Sci. USA 107, 11352–11357 (2010).
Sun, J. & MacKinnon, R. Cryo-em structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell 169, 1042–1050.e9 (2017).
Whicher, J. R. & MacKinnon, R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Sci. (80-.) 353, 664–669 (2016).
Wang, W. & MacKinnon, R. Cryo-em structure of the open human ether-à-go-go-related k + channel hERG. Cell 169, 422–430.e10 (2017).
Lee, C. H. & MacKinnon, R. Structures of the human HCN1 hyperpolarization-activated channel. Cell 168, 111–120.e11 (2017).
Tao, X., Hite, R. K. & MacKinnon, R. Cryo-EM structure of the open high-conductance Ca2+ -activated K+ channel. Nature 541, 46–51 (2017).
Tao, X. & Mackinnon, R. Cryo-EM structure of the KvAP channel reveals a non-domain-swapped voltage sensor topology. Elife 8, 1–15 (2019).
Clark, M. D., Contreras, G. F., Shen, R. & Perozo, E. Electromechanical coupling in the hyperpolarization-activated K + channel KAT1. Nature 583, 145–149 (2020).
Warmke, J. W. & Ganetzky, B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc. Natl Acad. Sci. USA 91, 3438–3442 (1994).
Sanguinetti, M. C., Jiang, C., Curran, M. E. & Keating, M. T. A mechanistic link between an inherited and an acquird cardiac arrthytmia: HERG encodes the IKr potassium channel. Cell 81, 299–307 (1995).
Curran, M. E. et al. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 80, 795–803 (1995).
Vandenberg, J. I. et al. hERG K(+) channels: structure, function, and clinical significance. Physiol. Rev. 92, 1393–1478 (2012).
Sanguinetti, M. C. & Tristani-Firouzi, M. hERG potassium channels and cardiac arrhythmia. Nature 440, 463–469 (2006).
Trudeau, M. C., Warmke, J. W., Ganetzky, B. & Robertson, G. A. HERG, a human inward rectifier in the voltage-gated potassium channel family. Sci. (80-.) 269, 92–95 (1995).
Schönherr, R. & Heinemann, S. H. Molecular determinants for activation and inactivation of HERG, a human inward rectifier potassium channel. J. Physiol. 493, 635–642 (1996).
Smith, P. L., Baukrowitz, T. & Yellen, G. The inward rectification mechanism of the HERG cardiac potassium channel. Nature 379, 833–836 (1996).
Spector, P. S., Curran, M. E., Zou, A., Keating, M. T. & Sanguinetti, M. C. Fast inactivation causes rectification of the IKr channel. J. Gen. Physiol. 107, 611–619 (1996).
Ferrer, T. et al. Molecular coupling in the human ether-a-go-go-related gene-1 (herg1) k+ channel inactivation pathway. J. Biol. Chem. 286, 39091–39099 (2011).
Li, J., Shen, R., Reddy, B., Perozo, E. & Roux, B. Mechanism of C-type inactivation in the hERG potassium channel. Sci Adv. 7, 1–8 (2021).
Lörinczi, É. et al. Voltage-dependent gating of KCNH potassium channels lacking a covalent link between voltage-sensing and pore domains. Nat. Commun. 6, 6672 (2015).
Butler, A., Helliwell, M. V., Zhang, Y., Hancox, J. C. & Dempsey, C. E. An update on the structure of HERG. Front. Pharmacol. 10, 1–13 (2020).
Costa, F., Guardiani, C. & Giacomello, A. Molecular dynamics simulations suggest possible activation and deactivation pathways in the hERG channel. Commun. Biol. 5, 1–11 (2022).
Shao, C. et al. Electrophysiological study of V535M hERG mutation of LQT2. J. Huazhong Univ. Sci. Technol. —Med. Sci. 31, 741–748 (2011).
Gustina, A. S. & Trudeau, M. C. The eag domain regulates hERG channel inactivation gating via a direct interaction. J. Gen. Physiol. 141, 229–241 (2013).
Perry, M. D., Ng, C. A. & Vandenberg, J. I. Pore helices play a dynamic role as integrators of domain motion during Kv11.1 channel inactivation gating. J. Biol. Chem. 288, 11482–11491 (2013).
Wang, D. T., Hill, A. P., Mann, S. A., Tan, P. S. & Vandenberg, J. I. Mapping the sequence of conformational changes underlying selectivity filter gating in the Kv 11.1 potassium channel. Nat. Struct. Mol. Biol. 18, 35–42 (2011).
Phan, K. et al. The S1 helix critically regulates the finely tuned gating of Kv11.1 channels. J. Biol. Chem. 292, 7688–7705 (2017).
Codding, S. J. & Trudeau, M. C. The hERG potassium channel intrinsic ligand regulates N- and C-terminal interactions and channel closure. J. Gen. Physiol. 151, 478–488 (2019).
Codding, S. J., Johnson, A. A. & Trudeau, M. C. Gating and regulation of KCNH (ERG, EAG, and ELK) channels by intracellular domains. Channels 14, 294–309 (2020).
Guo, J. & Zhou, H. X. Protein allostery and conformational dynamics. Chem. Rev. 116, 6503–6515 (2016).
Elbahnsi, A. & Delemotte, L. Structure and sequence-based computational approaches to allosteric signal transduction: application to electromechanical coupling in voltage-gated ion channels: computational approaches to allostery in ion channels. J. Mol. Biol. 433, 167095 (2021).
Guardiani, C. et al. Computational methods and theory for ion channel research. Adv. Phys. X 7, 2080587 (2022).
Beckstein, O. et al. Ion channel gating: Insights via molecular simulations. FEBS Lett. 555, 85–90 (2003).
Bassetto, C. A. Z., Carvalho-De-souza, J. L. & Bezanilla, F. Molecular basis for functional connectivity between the voltage sensor and the selectivity filter gate in shaker k+ channels. Elife 10, 1–30 (2021).
Carvalho-de-Souza, J. L. & Bezanilla, F. Noncanonical mechanism of voltage sensor coupling to pore revealed by tandem dimers of Shaker. Nat. Commun. 10, 3584 (2019).
Fernández-Mariño, A. I., Harpole, T. J., Oelstrom, K., Delemotte, L. & Chanda, B. Gating interaction maps reveal a noncanonical electromechanical coupling mode in the Shaker K+ channel. Nat. Struct. Mol. Biol. 25, 320–326 (2018).
Dijkstra, E. W. A note on two problems in connexion with graphs. Numer. Math. 1, 269–271 (1959).
de la Peña, P., Domínguez, P. & Barros, F. Gating mechanism of Kv11.1 (hERG) K+ channels without covalent connection between voltage sensor and pore domains. Pflug. Arch. Eur. J. Physiol. 470, 517–536 (2018).
Brandes, U. A faster algorithm for betweenness centrality. J. Math. Sociol. 25, 163–177 (2001).
Ju, P. et al. The pore domain outer helix contributes to both activation and inactivation of the hERG K+ channel. J. Biol. Chem. 284, 1000–1006 (2009).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Vardanyan, V. & Pongs, O. Coupling of voltage-sensors to the channel pore: A comparative view. Front. Pharmacol. 3 JUL, 1–10 (2012).
Alonso-Ron, C., De La Peña, P., Miranda, P., Domínguez, P. & Barros, F. Thermodynamic and kinetic properties of amino-terminal and S4-S5 loop HERG channel mutants under steady-state conditions. Biophys. J. 94, 3893–3911 (2008).
Hardman, R. M., Stansfeld, P. J., Dalibalta, S., Sutcliffe, M. J. & Mitcheson, J. S. Activation gating of hERG potassium channels: S6 glycines are not required as gating hinges. J. Biol. Chem. 282, 31972–31981 (2007).
Ferrer, T., Rupp, J., Piper, D. R. & Tristani-Firouzi, M. The S4-S5 linker directly couples voltage sensor movement to the activation gate in the human ether-a’-go-go-related gene (hERG) K+ channel. J. Biol. Chem. 281, 12858–12864 (2006).
Labro, A. J. & Snyders, D. J. Being flexible: The voltage-controllable activation gate of Kv channels. Front. Pharmacol. 3 SEP, 1–12 (2012).
Chowdhury, S. & Chanda, B. Thermodynamics of electromechanical coupling in voltage-gated ion channels. J. Gen. Physiol. 140, 613–623 (2012).
Choveau, F. S. et al. Opposite effects of the S4-S5 linker and PIP2 on voltage-gated channel function: KCNQ1/KCNE1 and other channels. Front. Pharmacol. 3 JUL, 1–16 (2012).
Patlak, J. B. Cooperating to unlock the voltage-dependent K channel. J. Gen. Physiol. 113, 385–387 (1999).
Bett, G. C. L., Zhou, Q. & Rasmusson, R. L. Models of HERG gating. Biophys. J. 101, 631–642 (2011).
Lees-Miller, J. P. et al. Interactions of H562 in the S5 Helix with T618 and S621 in the pore helix are important determinants of herg1 potassium channel structure and function. Biophys. J. 96, 3600–3610 (2009).
Zhang, M., Liu, J. & Tseng, G. N. Gating charges in the activation and inactivation processes of the hERG channel. J. Gen. Physiol. 124, 703–718 (2004).
Zagotta, W. N., Hoshl, T., Dittman, J. & Aldrich, R. W. Shaker potassium channel gating II: Transitions in the activation pathway. J. Gen. Physiol. 103, 279–319 (1994).
Liu, S., Rasmusson, R. L., Campbell, D. L., Wang, S. & Strauss, H. C. Activation and inactivation kinetics of an E-4031-sensitive current from single ferret atrial myocytes. Biophys. J. 70, 2704–2715 (1996).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Dickson, C. J. et al. Lipid14: The amber lipid force field. J. Chem. Theory Comput. 10, 865–879 (2014).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Bondy, J. A. & Murty, U. S. R. Graph theory with applications. (Elsevier Science Publishing Co., Inc., 1976).
Kraskov, A., Stögbauer, H. & Grassberger, P. Estimating mutual information. Phys. Rev. E—Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 69, 16 (2004).
Hagberg, A. A., Schult, D. A. & Swart, P. J. Exploring network structure, dynamics, and function using NetworkX. in 7th Python in Science Conference (SciPy 2008) 11–15 (2008).
Janke, W. Statistical Analysis of Simulations: Data Correlations and Error Estimation. in Proceedings of the Euro Winter School Quantum Simulations of Complex Many-Body Systems: From Theory to Algorithms Vol. 10 422–445 (2002).
Frenkel, D. & Smit, B. Understanding molecular simulation from algorithms to applications. (Academic Press, 2001).
Stefani, E. & Bezanilla, F. Cut-open oocyte voltage-clamp technique. Methods Enzymol. 293, 300–318 (1998).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
This research is part of the project that has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 803213) (F.C., C.G., and A.G.). F.C., C.G., and A.G. acknowledge PRACE for awarding us access to Marconi100 at CINECA, Italy. This work was also supported by National Institutes of Health (NIH, United States) Award R01GM030376 (C.B. and F.B.).
Author information
Authors and Affiliations
Contributions
C.B. (experiments) and F.C. (simulations) performed the research and wrote the paper. C.B., C.G., A.G. and F.B. designed the research and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mounir Tarek and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bassetto, C.A.Z., Costa, F., Guardiani, C. et al. Noncanonical electromechanical coupling paths in cardiac hERG potassium channel. Nat Commun 14, 1110 (2023). https://doi.org/10.1038/s41467-023-36730-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-36730-7
This article is cited by
-
An LQT2-related mutation in the voltage-sensing domain is involved in switching the gating polarity of hERG
BMC Biology (2024)
-
Hydrophobically gated memristive nanopores for neuromorphic applications
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.