Abstract
N6-methyladenosine (m6A) has been demonstrated to regulate RNA metabolism and various biological processes, including gametogenesis and embryogenesis. However, the landscape and function of m6A at single cell resolution have not been extensively studied in mammalian oocytes or during pre-implantation. In this study, we developed a single-cell m6A sequencing (scm6A-seq) method to simultaneously profile the m6A methylome and transcriptome in single oocytes/blastomeres of cleavage-stage embryos. We found that m6A deficiency leads to aberrant RNA clearance and consequent low quality of Mettl3Gdf9 conditional knockout (cKO) oocytes. We further revealed that m6A regulates the translation and stability of modified RNAs in metaphase II (MII) oocytes and during oocyte-to-embryo transition, respectively. Moreover, we observed m6A-dependent asymmetries in the epi-transcriptome between the blastomeres of two-cell embryo. scm6A-seq thus allows in-depth investigation into m6A characteristics and functions, and the findings provide invaluable single-cell resolution resources for delineating the underlying mechanism for gametogenesis and early embryonic development.
Similar content being viewed by others
Introduction
N6-methyladenosine (m6A) is one of the most important RNA modifications for the eukaryotic transcriptome1. m6A regulates mRNA alternative splicing2, export3, genome accessibility4, and transposon activity5,6 in nucleus, and also affects mRNA translation efficiency, degradation, and stability7 in cytoplasm. m6A is present along the whole transcriptome, and mainly enriched in coding region and 3’ UTR on the mRNAs8,9,10,11,12. Thus, m6A has been widely studied to exert extensive regulatory functions in gametogenesis and embryonic developmental events13.
Many studies have highlighted the roles played by m6A during oocyte development. In zebrafish, the m6A modified maternal transcripts tend to be recognized by Ythdf2 and degraded faster than the unmethylated transcripts14. In mice, Mettl3 deficiency led to a highly sterile phenotype in females, with features of abnormal oocyte ovulation and disordered oocyte maturation15. Ythdf2 depletion resulted in female-specific infertility and maternal transcript dosage during meiotic maturation16. The m6A methyltransferase complex component KIAA1429 is highly expressed in germinal vesicle (GV) oocytes and Kiaa1429 depletion in female mice showed small ovaries, accompanied by significantly decreased numbers of surrounded nucleolus (SN) oocytes and abnormal oocytes competence17. Moreover, in the cleavage-stage embryos, maternal Mettl3 depleted zygotes were arrested at the 1-cell stage15. Zygotic Mettl3 knockdown led to the failure of zygotic genome activation (ZGA), and the decreased m6A abundance from the GV oocytes to the 2-cell embryos18.
ZGA of the mice zygote embryos begins at the late 1-cell embryos, and many studies have pointed to the significance of chromatin remodeling in zygotic transcription initiation, including DNA methylation, histone modifications, chromatin accessibility, high-order chromatin structures formation, and RNA polymerase II (Pol II) binding19,20,21,22. Interestingly, in addition to the newly synthesized zygotic transcripts, ZGA-dependent maternal RNA clearance processes are crucial for early embryonic development23,24. Moreover, RNA metabolism during ZGA results in the heterogeneity of epigenomic modification among blastomeres and suggests transcriptome-wide regulation initiated at the 2-cell embryonic stage25,26.
Previous studies have depicted the landscapes of the single-cell transcriptome27,28,29, and low input-based translatome30,31 and m6A methylome32 in oocyte and early stages of embryos. But the characteristics and functions of RNA methylation at single-cell level remain unclear. Accumulated methods, which are mainly based on antibody enrichment, sensitive enzymatic digestion or chemical conversion, have been developed to profile and characterize transcriptome-wide m6A from bulk to low-input samples8,32,33,34,35,36,37. Recently, scDART-seq38 using exogenously expressed APOBEC1-YTH construct showed the power of single-cell m6A detection, opening a new avenue for investigating epitranscriptomic regulation at the single-cell level.
In this study, we developed a single-cell m6A sequencing (scm6A-seq) method without exogenous gene expression. The scm6A-seq combines the principle of RNA multiplex labeling technology39 with methylated RNA immunoprecipitation sequencing (MeRIP-seq/m6A-IP), enabling the capture of the transcriptome-wide m6A landscape and comparison of the m6A level among single cells. Utilizing scm6A-seq, we interrogate the m6A modification of mouse oocytes and early embryos. Our findings revealed dynamic m6A regulation in RNA metabolism at single-cell resolution during oocyte development and ZGA.
Results
Establishment of the single-cell m6A sequencing method
To profile the whole RNA m6A modified transcriptome in single cells, we developed the single-cell m6A sequencing (scm6A-seq) method, in which the RNAs from each cell were first fragmented and ligated, and then labeled with two rounds of barcoded DNA adapters for parallelized single-cell sequencing. The first round of labeling involved ligating the adapters with barcode 1 to the fragmented RNAs from each cell, followed by the second round of labeling to add the barcode 2 adapters to the 3’ end of barcode 1 adapters through base-paring (Supplementary Fig. 1a). Then, the barcoded RNAs from different cells were pooled together and subjected to RNA-seq and m6A immunoprecipitation (m6A-IP), respectively (“Methods”). During library construction, a dSpacer-blocked template switch oligo (TSO) and T7 primer were used for cDNA synthesis and amplification, and then, T7 was used for in vitro transcription, ribosomal RNA (rRNA) depletion, and PCR amplification and sequencing (Fig. 1a and Supplementary Fig. 1a). Overall, scm6A-seq enables simultaneous transcriptome and m6A methylome sequencing at single-cell resolution through specific barcode labeling.
a Schematic diagram showing the scm6A-seq method and an overview of the samples analyzed. The carton image of oocytes and early embryos is created with BioRender. b Saturation curves for each GV oocyte data obtained by scm6A-seq. Each point on the curve was generated by randomly selecting a number of raw reads from each GV cell library and then using the same alignment pipeline to call covered genes. c Bar plot showing the number of detected m6A peaks and modified transcripts in each GV oocyte using scm6A-seq. d The distribution of mRNA m6A peaks in individual GV oocytes. e Metagene profiles depicting the normalized sequence coverage ratio of m6A-IP to RNA-seq data in positions surrounding the transcription start site (TSS) (left) and translation stop codon (right). The red line and light red shadow represent the mean and quantiles of 10 GV oocytes determined by scm6A-seq, respectively. The other three colored lines represent bulk MeRIP-seq and RNA-seq, as labeled in the panel. f Bar plot displaying the accumulative fraction of the number of the commonly m6A modified transcripts in 10 GV oocytes. Source data are provided as a Source data file. g The box and point plot showing the expression of common m6A modified transcripts. The x-axis shows the number of common m6A peaks in 10 GV oocytes. Source data are provided as a Source data file. h Venn diagram displaying the common m6A modified transcripts between single cells and bulk cells detected from MeRIP-seq data. Light green pie represents identified modified RNAs identified in 70 GV oocytes by MeRIP-seq, and the purple pies represent the common m6A modified RNAs form 10 individual GV oocytes as determined by scm6A-seq. n represents the frequency with which common m6A modified RNAs appeared in the analysis of the 10 individual GV oocytes.
To determine whether scm6A-seq can be used to identify m6A peaks in single cells, we first performed scm6A-seq on germinal vesicle (GV) oocytes. Overall, we obtained an average of 12 million mapped reads and more than 15,000 detected RNAs in each of 10 GV oocytes (Fig. 1b). High expression correlation was observed among 10 individual GV oocytes, especially in the RNA-seq data (Supplementary Fig. 1b), justifying the reproducibility and sensitivity of scm6A-seq. Attributing to the sufficient sequencing depth and coverage, we further identified high-confident m6A features (“Methods”), and obtained more than 5000 m6A modified peaks in over 4000 RNAs in each oocyte (Fig. 1c and Supplementary Data 1). To validate the accuracy and specificity of the detected m6A peaks by scm6A-seq, we also performed RNA-seq and methylated RNA immunoprecipitation (MeRIP-seq) using tens of oocytes (25, 50, and 70) and used the same criteria for filtering the peaks (Supplementary Data 2). More than 80% of m6A modified RNAs obtained by both single-cell and bulk sequencing methods were protein-coding genes, and a slightly higher proportion in scm6A-seq (Supplementary Fig. 1c), and the modified peaks were mainly located in their coding sequence (CDS), stop codon regions and 3’UTRs (Fig. 1d). Metagene analysis of the identified m6A peaks in oocytes showed an obvious enrichment of m6A signals around transcription start sites (TSS) and stop codon regions (Fig. 1e), and conserved RRAC motif (R = A/G) was observed in high-confidence m6A peaks (Supplementary Fig. 1d), which are consistent with previous reports15.
We further characterized the m6A features in single cell identified by scm6A-seq. First, we detected a good repeatability of m6A modified RNAs identified from tens of GV oocytes using the same method of scm6A-seq (Supplementary Fig. 1e). We then counted the m6A modified RNAs among 10 individual GV oocytes and found that over 40% m6A modified RNAs can be identified in more than 5 oocytes (Fig. 1f). Moreover, we also observed that the expression level of m6A modified RNAs was increased along with their frequencies in 10 individual oocytes (Fig. 1g). Comparing the modified RNAs in single cell with those in tens of oocytes sample, we found that the conserved modified RNAs with high frequencies was more likely to be detected in 70 GVs samples, such as 1 out of 10 oocytes in scm6A-seq with 40.96%, 4/10 with 78.46%, 6/10 with 87.94%, and 10/10 with 97.60% (Fig. 1h). Furthermore, we defined the commonly modified RNAs as those with m6A peak enrichment scores in the top 5000 for each oocyte and meanwhile, identified in no less than 6 oocytes. A total of 1816 commonly modified RNAs were categorized into this group accounting for more than 60% of m6A modified RNAs in each oocyte, in relative to only around 40% in tens of oocytes sample (Supplementary Fig. 1f). Importantly, nearly 70% of the modified RNAs identified by bulk MeRIP-seq15 and 55% by low input sequencing (ULI-MeRIP-seq)32 of GV oocytes were also detected in merged single GV oocyte by scm6A-seq (Supplementary Fig. 1 g). These commonly modified RNAs identified by three detection techniques were mainly involved in cell cycle, DNA repair, and cell division pathways (Supplementary Fig. 1h), such as E2f1 and Bod1 (Supplementary Fig. 1i). Notably, some modified RNAs were identified in most proportion of cells only by scm6A-seq, but not by low input and bulk sequencing, such as the genes of Hist4h4, Hist1h4f, and Mapk3 involving cell cycle and DNA replication (Supplementary Fig. 1i). Thus, scm6A-seq enables robust m6A identification in single cells and is more sensitive especially for low-abundant modified RNAs.
METTL3 promotes degradation of methylated RNAs in GV oocytes
In Mettl3Gdf9 conditional knockout (cKO) mice (Mettl3flox/flox;Gdf9-cre), the ovary size was largely decreased and meanwhile the oocyte maturation was also blocked. Over 80% of the oocytes were arrested at GVBD stage and the percentage of MII oocytes was less than 5%. Moreover, zygotes derived from Mettl3Gdf9 cKO oocytes were arrested at 1-cell stage after fertilization15. But whether this phenotype is mediated by METTL3-dependent m6A in oocytes remained to be determined. The scm6A-seq was then performed on control and Mettl3Gdf9 cKO oocytes in the GV stage to identify METTL3-dependent modified RNAs. We observed globally decreased m6A signals in Mettl3Gdf9 cKO oocytes (Supplementary Fig. 2a), especially near stop codons (Fig. 2a). A total of 1214 RNAs, including over 5% modified ncRNAs (Supplementary Fig. 2b), had lost m6A signatures in Mettl3Gdf9 cKO GV oocytes, which were defined as the METTL3-dependent m6A modified RNAs (Fig. 2b, Supplementary Data 3). These RNAs in control oocytes showed m6A peaks embedded in conserved RRACH motif sequences (R = A/G, H = A/C/G) (Supplementary Fig. 2c) and significantly enriched in mitotic cell cycle and cell division processes (Fig. 2c), indicating that the cell cycle related modified RNAs are probably implicated in oocytes maturation. To further investigate the role of m6A in regulating RNA metabolism in oocytes, we compared the expression changes of METTL3-dependent m6A-modified RNAs with the unmodified or modified ones independent of METTL3 in Mettl3Gdf9 cKO versus the control oocytes. The results showed that the expression of METTL3-dependent m6A-modified RNAs in Mettl3Gdf9 cKO oocytes is significantly enhanced in relative to the unmodified RNAs (Fig. 2d), suggesting that METTL3-dependent m6A-modified RNAs are preferentially degraded in control GV oocytes, but stabilized in Mettl3Gdf9 cKO oocytes owing to the loss of m6A modification. Intriguingly, significantly increased expression of METTL3-independent m6A-modified RNAs relative to the unmodified RNAs was also observed (Fig. 2d). The RNA abundance of m6A modified RNAs is higher than the unmodified ones (Supplementary Fig. 2d), which is consistent with the previous report in single cell38 and may be the reason for the high expression of METTL3-independent m6A-modified RNAs in Mettl3 cKO oocytes. Since YTHDF2 has been identified as a m6A reader protein mediating the degradation of its targeted m6A modified RNAs16,40,41, and substantially higher translation signal30 was observed in the fully-grown oocytes (Fig. 2e), we further analyzed the YTHDF2 binding sites from the public YTHDF2 enhanced crosslinking and immunoprecipitation sequencing (eCLIP-seq) data previously generated from mouse embryonic stem cells (mESCs)42 and found that METTL3-dependent m6A signatures significantly overlap with the YTHDF2 targets compared with the random background (Fig. 2f). Furthermore, we identified significantly upregulated RNA expression upon Mettl3Gdf9 cKO within the METLL3-dependent m6A modified RNA populations (Supplementary Fig. 2e), and found that these RNAs, such as Cdc25b, Bag6, B4galt5, Strip1, and Smn1, were involved in cell cycle and metabolism processes (Fig. 2g and Supplementary Fig. 2f). Collectively, our results suggested that METTL3-catalyzed m6A mainly regulates RNA stability and preferentially promotes degradation in GV oocytes.
a Metagene profiles depicting m6A signals surrounding the stop codon in control and Mettl3 conditional knockout (cKO) GV oocytes. b Heatmap showing the METTL3-dependent m6A modified transcripts in GV oocytes. c Functional annotation analysis of METTL3-dependent m6A modified transcripts using Metascape. The enrichment and P value was calculated with default parameters using hypergeometric test of functional annotation in DAVID database. d Cumulative frequency of expression change (log2(fold change)) of METTL3-dependent, METTL3-independent m6A modified and unmodified RNAs upon Mettl3 silencing. P values were determined by one-sided Wilcoxon test. P = 1.2e−10, between METTL3-independent and W/O m6A RNA sets. P < 2.2e−16, between METTL3-dependent and W/O m6A RNA sets. ***P < 0.001. e Integrated Genomics Viewer (IGV) diagram displaying the translation signals of Ythdf2 in oocytes and zygotes. f Density plot displaying the distance between YTHDF2 target peaks detected by enhanced crosslinking and immunoprecipitation sequencing (eCLIP-seq) in mESCs42 (GSE151788) and METTL3-dependent m6A peaks in GV oocytes by scm6A-seq. g Integrated Genomics Viewer (IGV) diagram showing the METTL3-dependent m6A peaks across Cdc25b, Bag6, and B4galt5. The light blue boxes represent the identified m6A peaks identified by scm6A-seq.
scm6A-seq distinguishes SN and NSN GV oocytes
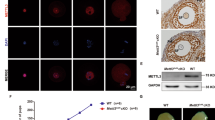
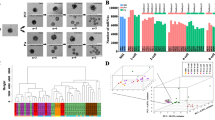
The successful identification of m6A modification in single cells by scm6A-seq allows us to further classify the oocytes populations (30 control and 27 Mettl3Gdf9 cKO GV oocytes) using m6A signatures. Through scm6A-seq, we not only barcoded individual cells prior to m6A enrichment, but also pooled the barcoded cells together in the same batch for library construction and sequencing, which allowed direct comparison of the m6A levels among individual cells using the data obtained from the same batch. In addition, scm6A-seq enables joint profiling of m6A methylome and transcriptome in individual cells, producing RNA-seq, m6A-IP and supernatant data for each cell (Supplementary Fig. 1a). We found that the control GV and Mettl3Gdf9 cKO GV oocytes can be distinguished by using each expression matrix of RNA-seq, m6A-IP and supernatant or their merged expression matrix from scm6A-seq data in single cells (Fig. 3a–c and Supplementary Fig. 3a). Meanwhile, further including the single-cell m6A level in the matrix improved the accuracy in distinguishing the control and Mettl3Gdf9 cKO GV oocytes due to a decreased m6A level in the Mettl3Gdf9 cKO oocytes (Fig. 3d). Intriguingly, the cells were clustered into three classes when only transcriptome data were used (Supplementary Fig. 3b), but we observed two sub-populations in the control and Mettl3Gdf9 cKO oocyte groups when taking the m6A level of individual oocytes as a third component for clustering (Fig. 3e and Supplementary Fig. 3c). Previous study reported that there are both SN and NSN GV oocytes on the basis of their growth-related transcription in the ovary of the mice after ovulation induction, with the NSN oocytes displaying lower developmental capacity43. Thus, the sub-populations in control and Mettl3Gdf9 cKO groups were considered to correspond to SN and NSN oocytes which were further validated by the marker genes of Gata3 and Mapk7 for oocyte growth44, respectively (Fig. 3f). Meanwhile, we also observed a lower RNA abundance in NSN oocytes, corresponding to their transcription activity and incomplete growth (Fig. 3g). We also found a higher proportion of NSN oocytes in Mettl3Gdf9 cKO group (Supplementary Fig. 3d). And we also verified the existence of SN and NSN oocytes in the cumulus cell-oocyte complex (COC) by nucleus staining of both control and Mettl3Gdf9 cKO mice (Fig. 3h). Thus, the multi-omics data from scm6A-seq enables more precise classification of cell populations. To further clarify the differences between SN and NSN oocytes, we analyzed the most significantly differentially expressed RNAs between SN and NSN oocytes, and found that most of these RNAs were related to oocyte growth (Fig. 3i). On the other hand, the dynamic expression of m6A methyltransferases and readers during oocyte growth also suggested a m6A related mechanism of RNA metabolism during oocyte growth45 (Supplementary Fig. 3e). As trimethylated histone 3 lysine 36 (H3K36me3) has been reported to be involved in regulating gene expression during oocyte development46, we defined the decreased genes (log2fragments per kilobase of transcript per million mapped reads (FPKM) (fully-grown oocyte (FGO)/postnatal day 7 (P7)) < −2)) which lost the H3K36me3 during oocyte growth as oocyte development-silencing genes (Supplementary Fig. 3f). Because of the m6A-mediated RNAs degradation in GV oocytes (Fig. 2), we then evaluated the stability of oocyte development-silencing RNAs by measuring altered expression in control and Mettl3Gdf9 cKO SN GV oocytes and observed abnormal accumulation of development-silencing RNAs in Mettl3Gdf9 cKO oocytes (Supplementary Fig. 3g, h). These results suggest that the failure of degradation of development-silencing RNAs might be the crucial reason for the low quality of Mettl3Gdf9 cKO GV oocytes.
a–c Uniform manifold approximation and projection (UMAP) plot based on the reads count matrix of RNA-seq (a) and m6A-IP (b), and merged total reads count of single GV oocytes (c). The UMAP plot is colored on the basis of the cells type. Source data are provided as a Source data file. d The 3D plot showing the relative m6A level of each GV oocyte based on the UMAP results, colored by cells type (left) and cells reclassified by the comprehensive analysis of the gene expression matrix reduced dimensions by UMAP and m6A level (right). Source data are provided as a Source data file. e The two-dimensional UMAP diagram showing the cell populations on the basis of the classification results in (d). The Clusters 1–4 of cells were redefined as Control SN, Control NSN, Mettl3 cKO SN, and Mettl3 cKO NSN clusters according to the expression of marker RNAs and genome type of oocytes. Source data are provided as a Source data file. f Violin plot showing the quality of markers Gata3 and Mpak7 expression in oocytes in different clustering populations. g Combined box and scatter plot showing the difference in RNA amount between SN and NSN oocytes. n (Control SN) = 19, n (Control NSN) = 12, n (Mettl3 cKO SN) = 14, and n (Mettl3 cKO) = 12. The middle lines of the boxes represent the medians of datasets. The upper and bottom lines of the boxes are respectively the upper quantile and the lower quantile of the data. The whiskers mark the upper and lower limits of these datasets, respectively. The two-sided P value was calculated by unpaired student t test. Source data are provided as a Source data file. h Morphology of the pronuclear in both the SN and NSN oocytes isolated from the cumulus cell-oocyte complex (COC). Both control and METTL3-null oocytes were stained with DAPI and an anti-METTL3 antibody. n (Control) = 28, n (Mettl3Gdf9 cKO) = 14. The white rectangle represents the area of the nucleus to be enlarged. Source data are provided as a Source data file. i Heatmap showing the expression levels of presentative genes in both control and METTL3-null oocytes (left) and oocytes at different growth stages (right). The representative genes were identified by the most differentially expressed genes in the SN and NSN oocytes compared to control oocytes. Source data are provided as a Source data file.
The dynamic m6A methylome during oocyte maturation
The mammalian oocyte maturation process is coupled with maternal RNA translational activation and degradation47,48, including GV, germinal vesicle breakdown (GVBD) after the resumption of oocyte meiosis, and two consecutive metaphase (MI and MII) stages. Fully- grown GV oocytes are transcriptionally silent, making the oocytes maturation an ideal model for post-transcriptional regulation detection. To determine the regulation of m6A deposition during oocyte maturation, we performed scm6A-seq on cultured oocytes in the GV, GVBD, MI, and MII stages (10 oocytes for each stage) (Supplementary Data 4). We first summarized the changed RNAs expression during oocyte maturation using K-means clustering from RNA-seq of scm6A-seq, and obtained a cluster of degraded RNAs related to the cell cycle and oocyte maturation (Fig. 4a, b) and another set of RNAs with increased expression correlated with translation and protein transport (Supplementary Fig. 4a, b), which findings are consistent with the tendency of RNA metabolism during oocyte maturation. However, RNA expression matrix alone cannot accurately distinguish the stage-specific cells during oocyte maturation (Supplementary Fig. 4c). Therefore, we next depicted the m6A signatures of oocyte in different maturation stages by scm6A-seq (Supplementary Fig. 4d) and observed some stable m6A modified RNAs during oocytes maturation, such as Lars2 (Supplementary Fig. 4e), involving in the signaling pathways (Supplementary Fig. 4f). Additionally, we also observed many stage-specific m6A-modified RNAs, such as Bnc2, Tmed9, Arcn1, and Cd320 (Supplementary Fig. 4g), which were involved in differential functions, especially nucleosome assembly, transcription in GV oocytes, and spindle assembly, protein transport in M-phases oocytes (Supplementary Fig. 4h). These results suggest a non-co-transcriptionally regulatory mechanism of m6A during oocyte maturation. Interestingly, we observed an increased m6A enrichment during the oocyte maturation based on the metagene analysis and estimation of m6A level (Fig. 4c and Supplementary Fig. 4i). What’s more, oocytes of different stages were well clustered when m6A features were included (Fig. 4d). Because oocyte maturation is accompanied by maternal RNA translational activation, we assessed the translation differences and found the m6A modified transcripts tend to have significantly higher translation levels than the unmethylated transcripts (Fig. 4e). We then aimed to determine if this process is regulated by the m6A readers of YTHDF1 and YTHDF3, which were validated with capability of enhancing the translation of m6A modified transcripts49,50. Based on public proteomic data generated from oocyte maturation51, we observed a high level of YTHDF3 versus a low level of YTHDF1 in MII stage (Fig. 4f). In addition, we also observed considerate overlap between the m6A peaks and the YTHDF3 targets by eCLIP-seq of mESCs42 (Fig. 4g). Collectively, these results indicate that m6A promotes the translation of m6A modified transcripts in MII stage, and this effect is potentially mediated by YTHDF3.
a Line plot showing the decrease in expression of the gene set during oocyte maturation as indicated by K-means clustering. b Bar plot displaying the enriched Gene Ontology (GO) terms in the gene set showing in (a). The enrichment and P value were calculated with default parameters using hypergeometric test of functional annotation in DAVID database. c Metagene profiles depicting m6A signals in areas surrounding the transcription start site (TSS) (left) and stop codon (right). Colored lines represent different stages of oocyte maturation. d Uniform manifold approximation and projection (UMAP) clustering for oocytes at different stages using the merged omics data obtained by scm6A-seq. e Cumulative frequency of translation signals89 (GSE169632) for RNAs with or without m6A modification in MII oocytes. P values were determined by one-sided Wilcoxon test, P < 2.2e−16. ***P < 0.001. f Heatmap displaying the protein abundance of YTHDF1/2/3 in GV and MII oocytes. g Density plot displaying the distance between YTHDF3 target peaks detected by enhanced crosslinking and immunoprecipitation sequencing (eCLIP-seq) in mouse embryonic stem cells (mESCs)42 (GSE151788) and m6A modified RNAs in MII oocytes by scm6A-seq.
m6A protects maternal RNAs during MII-to-zygote transition
To clarify the regulatory role of m6A during oocyte-to-embryo transition, we performed scm6A-seq on different stages of embryos: zygote (1 C), early 2-cell (E2C), mid-2-cell (M2C), late 2-cell (L2C), and 4-cell (4C). Based on the transcriptome sequencing data, we observed 3 major clusters of cells from MII to 4C stage (Supplementary Fig 5a). We further identified three RNAs clusters based on the deferentially expressed RNAs during the developmental stages, defined as maternal decay, minor ZGA, and major ZGA clusters according to their expression changes (Fig. 5a and Supplementary Data 5), which are mainly involved in translation, cell division and embryonic cleavage pathways, respectively (Supplementary Fig. 5b–d). Of note, only less than 4% of maternal decay RNAs were marked by m6A in MII oocytes (Supplementary Fig. 5e). Intriguingly, from MII oocytes to early embryos, m6A modified RNAs had a significantly increased expression (Fig. 5b), whereas the unmodified ones showed a decreased expression (Supplementary Fig. 5f). Thus, the expression of m6A modified RNAs, but not the unmodified ones, tends to be increased from MII to zygote (Fig. 5c).
a Heatmap showing the normalized expression of significantly dysregulated RNAs between two adjacent time points during early embryo development. The expression of maternal decay genes is the upregulated in MII oocytes, that of minor zygotic genome activation (ZGA) genes is upregulated in the early and mid-2-cell stages, and that of major ZGA genes is upregulated in the late 2-cell and 4-cell stages. b Box and scatter plot showing the expression of m6A-modified RNAs in oocytes from the MII to the 4-cell stage. The middle lines of the boxes represent the medians of datasets. The upper and bottom lines of the boxes are respectively the upper quantile and the lower quantile of the data. The whiskers mark the upper and lower limits of these datasets respectively. P values were determined by one-sided Wilcoxon test, P = 2.611e−9. ***P < 0.001. c Cumulative fraction of RNA expression change (log2(fold change)) between zygote and MII oocytes of WT oocytes. P values were determined by one-sided Wilcoxon test, P < 2.2e−16. ***P < 0.001. d Cumulative fraction of RNA expression change (log2(fold change)) between zygotes and MII oocytes of Mettl3Gdf9 cKO oocytes. P values were determined by one-sided Wilcoxon test, P = 5.413e−11. ***P < 0.001. e Genome browser showing the representative RNA abundance of m6A modified maternal RNAs in control and Mettl3Gdf9 cKO MII oocytes and zygotes. f Heatmap showing the expression level of m6A-related RNAs in scm6A-seq data during early embryonic development. g Protein abundance of expressed m6A-related proteins during early embryonic development. h Density plot displaying the distance between IGF2BP2 target peaks detected by enhanced crosslinking and immunoprecipitation sequencing (eCLIP-seq) in human embryo steam cells (hESCs)52 (GSE78509) and m6A peaks identified in MII oocytes by scm6A-seq.
Intriguingly, the m6A-modified RNAs degrade more rapidly than the unmodified ones in early and late 2-cell stages (Supplementary Fig. 5g and Supplementary Data 6). These results support different mechanisms of m6A in regulating RNA metabolism in oocytes and early embryos, which has also been well discussed in the recent report32. To further confirm the metabolism of m6A-modified RNAs during MII to Zygote transition, we collected the normal control and Mettl3Gdf9 cKO MII oocytes and zygotes for total transcriptome sequencing (Supplementary Data 7). We found that in the Mettl3Gdf9 cKO samples, the RNA metabolism is largely disordered (Supplementary Fig. 5h) and the m6A modified RNAs in MII are rapidly degraded during Mettl3Gdf9 cKO MII-to-zygote transition (Fig. 5d). Besides, a higher proportion of the m6A modified RNAs in oocytes are enriched in the hypo-stable group (Supplementary Fig. 5i), but substantially decrease during MII-to-zygote transition in the Mettl3Gdf9 cKO samples (Fig. 5e). These results suggest that m6A protects RNA from degradation in an METTL3-dependent manner.
To investigate the RNA binding proteins for regulation, we further evaluated the transcription and protein abundance of the m6A-related writers, erasers, and readers during oocyte-to-embryo transition. We observed persistent transcription of Wtap, Alkbh5, and Igf2bp2 in the RNA-seq data of scm6A-seq (Fig. 5f). And IGF2BP2 protein abundance remains high at oocyte and zygote stages but decreases upon entry into the 2-cell stage (Fig. 5g, Supplementary Fig. 5j), and whose translation signal is decreased after fertilization (Supplementary Fig. 5k). Besides, we also observed considerable co-localization between m6A peaks of maternal RNAs detected in MII oocytes and the binding targets of IGF2BP2 in ESCs identified by eCLIP-seq52 (GSE78509) (Fig. 5h), suggesting that IGF2BP2, as a m6A reader protein, might recognize m6A modified maternal RNAs in the oocytes and enhance their stability during MII-to-zygote transition.
scm6A-seq reveals the heterogeneity of 2-cell blastomeres
To further explore the role played by m6A in early embryonic development, we characterized the m6A signatures at single blastomere resolution and observed a trend of decreased m6A level after ZGA (Supplementary Fig. 6a), which is consistent with the result detected by HPLC in the previous report32. And the m6A enriched fragments showed stage specificity but no significant correlation with RNA expression (Fig. 6a, Supplementary Fig. 6b and Supplementary Data 8), pointing to stage-specific functions (Supplementary Fig. 6c). More elaborate stage-specific cell populations were observed when we introduced m6A signatures into a clustering assay (Fig. 6b). These results suggest a higher sensitivity and reliability of scm6A-seq in identification of m6A-level and classification of single cells.
a Heatmap displaying the expression of m6A-modified RNAs during early embryonic development. b Uniform manifold approximation and projection (UMAP) clustering of cells during early embryonic development. c Principal component analysis (PCA) with cells in the early, mid-, and late 2-cell stages using merged omics data of scm6A-seq. Cells are colored by cluster population. Source data are provided as a Source Data file. d Annotated stage information for the analysis presented in (c). Cells are colored by stage information as indicated. e The proportion of cells in each cluster of different 2-cell stage oocytes. f The proportion of cells in each stage for different clustering populations. g PCA plot displaying the cell pairing information of early, mid-, and late 2-cell embryos. h Bar plot showing the proportion of pairing information for different stages of the 2-cell embryos in two cluster populations. i TF mRNAs with the most enrichment motifs identified from promoter and gene body peaks of the differentially expressed genes among Cluster1 blastomeres and Cluster2 blastomeres. Source data are provided as a Source Data file. j Bar plot of Gene Ontology (GO) term enrichment for the TFs with m6A modification in the 4-cell stage embryos. The enrichment and P value were calculated with default parameters using hypergeometric test of functional annotation in DAVID database. k Genome browser showing the read abundance for Pou5f1 in early embryo cells as indicated by the scm6A-seq data. The yellow rectangle boxes represent identified m6A peaks in 4-cell embryos. l TF-target network displaying the modified TF mRNAs with their targets in 4-cell stage embryos. Diamonds are the modified TF mRNAs, and the circular dots represent their targets. The size of the TF mRNAs represents the number of interacting pairs.
The blastomeres of 2-cell embryos have been reported to be molecular heterogeneous and contribute unequally to the differentiation of the inner cell mass (ICM) and trophectoderm (TE) during embryonic development25,53,54. Moreover, asynchronous development of porcine blastomeres in the 2-cell stage has been observed55,56. Considering the heterogeneity of blastomeres, we further explored the m6A roles played in asynchronous development of mouse 2-cell embryos, and obtained two cell populations on the basis of the m6A and expression signatures obtained by scm6A-seq (Fig. 6c and Supplementary Fig. 6d) with stage information (Fig. 6d). We found that nearly 95% of early 2-cell blastomeres were in Cluster 1 (Fig. 6e), and more than 98% of the blastomeres in Cluster 2 were from mid-to-late 2-cell embryos (Fig. 6f). These data suggested that cells in Cluster 1 and 2 were in the early stage as minor ZGA and advanced stages of 2-cell as major ZGA, respectively. Furthermore, we labeled the pairing information in the 2-cell stages blastomeres (Fig. 6g) and found that a reduced proportion of paired blastomeres appeared in the same cell population during 2-cell embryos development (Fig. 6h), indicating that the heterogeneity between the two blastomeres is increased in the 2-cell embryos before major ZGA. We then analyzed the leading and lagging blastomeres in 2-cell embryos due to asynchronous ZGA (Supplementary Fig. 6e), and further defined the blastomeres in mid-to-late 2-cell embryos embedded in Cluster 1 are the lagging population and those in Cluster 2 are the leading population. Through comparing the m6A characteristics in these two clusters, we found a larger variance in Cluster 1 even though the difference in global m6A level between these two groups was insignificant (Supplementary Fig. 6f), suggesting a potential role for m6A in gene activation regulation.
We further compared the deferentially expressed and m6A-modified RNAs related to ZGA between the 2 clusters, and observed more significant changes of m6A signals than RNA expression (fold change >4) (Supplementary Fig. 6g). To identify the upstream regulators of these changes, we downloaded ATAC-seq data57 to correlate chromatin accessibility with RNA expression and m6A signals, and identified the activated RNAs in Cluster 2 with chromatin accessibility as RNAs involved in major ZGA and RNAs in Cluster 1 that had lost accessibility as silenced maternal RNAs (Supplementary Fig. 6h, i). Further analysis showed that the most RNAs involved in the major ZGA were protein-coding RNAs, while the silenced maternal RNAs contained a larger proportion of non-coding RNAs (Supplementary Fig. 6j). Regarding that transcription factors (TFs) binding is required for the chromatin accessibility and transcription58, we identified enriched TFs on the basis of the accessible regions of genes differentially expressed during ZGA as described in Supplementary Fig. 6h. Importantly, we found a significantly differential m6A signal, but the expression level of these TF mRNAs between the two clusters did not show significant difference (Fig. 6i). These results suggest that m6A modification on TF mRNAs may be involved in the regulation of ZGA.
During further exploration, the involvement of most m6A-modified TF mRNAs during embryonic development were found to be stage-specific (Supplementary Fig. 6k). Of note, only the m6A-modified TF mRNAs in 4-cell stage were significantly enriched in the m6A modified and expressed RNA populations (FPKM > 0.5), as determined by hypergeometric test (Supplementary Fig. 6l). In addition, the m6A modified TF mRNAs in each stage were involved in differential pathways, for example, those in MII oocytes and zygotes were annotated into cell proliferation and gene expression processes (Supplementary Fig. 6m, n), while those at early and mid-2-cell stage were enriched in development and signaling pathways (Supplementary Fig. 6o, p). Intriguingly, m6A modified TF mRNAs in the 4-cell stage were significantly involved in differentiation pathways (Fig. 6j), such as Pou5f1 (Fig. 6k), which was found to be essential for early embryogenesis59. Moreover, we built an interaction network of m6A-modified TF mRNAs with their targets from TRRUST database, and found that the TF mRNAs modified by m6A at 4-cell stage can target some critical factors that related to lineage differentiation, such as Cdx2, Nanog, and Sox2 (Fig. 6l). These results indicate the involvement of m6A in lineage specification through TFs.
Discussion
In this study, we developed a robust scm6A-seq method, which is simple, practical, and versatile tool to profile the whole transcriptomic m6A in single cells. In this study, the individual cells are labeled through two rounds of barcoding. Then, samples were pooled together for m6A enrichment using anti-m6A antibody IP and subject to library construction and sequenced parallelly. This strategy minimizes the batch effect and variance of enrichment efficiency among individual cells, enabling us to compare the relative m6A level directly among single cells. Optionally, a third-round labeling can also be performed during cDNA synthesis with barcoded TSO primers, which would allow flexible choice of labeling strategies as indicated by the sample. Recently, scDART-seq38 based on the expression of exogenously engineered YTH-APOBEC1 was developed as the first single-cell level detection method for m6A profiling. The advantage of our scm6A-seq is to detect the m6A at single-cell resolution under native conditions, which provides an additional powerful tool to tackle complex biological mechanisms underlying epitranscriptomic regulation.
Currently, many single-cell multi-omics technologies were reported to detect transcriptome, genome, or epigenomic markers at the same time in single cells60, and these tools are often used to study the regulation of gene expression by epigenomic factors. Therefore, sequencing technologies that combine the transcriptome and RNA modifications are highly desired to uncover the regulation of RNA metabolism by epitranscriptomic factors. Developing multi-omics technologies to profile transcriptome with multiple RNA methylome or translatome within one cell at the same time will be very intriguing. In addition, due to the sparsity of scm6A-seq data, there are some technical difficulties in the analysis of single-cell multi-omics data compared to bulk MeRIP-seq data. Therefore, data sparsity and multi-dimensional data integration analysis should be considered in the development of future algorithms and computing models.
Though there are many studies focusing on the functions of m6A methylome during oocyte and early embryo development, some scientific questions need to be further addressed: Firstly, only the m6A methylome of the fully-grown oocytes is available, and the mechanism of m6A in regulating maternal RNA metabolism during oocyte growth is still required to be evaluated, especially for the appearance of METTL3-independent m6A modified RNAs which may be catalyzed by other methyltransferase such as METTL1661. It will be very interesting to figure out the details of newly occurred m6A in Mettl3Gdf9 cKO oocytes and the different molecular fate of m6A-modified RNAs during oocyte development. Secondly, our results support the conclusion that m6A protects the m6A-modified maternal RNAs from degradation during the MII-to-zygote transition (Fig. 5b–e). However, we also found many hyper-stable transcripts in the Mettl3Gdf9 cKO oocytes and zygotes (Supplementary Fig. 5h), which may be caused by the decreased activity of RNA degradation machinery under the Mettl3Gdf9 cKO condition. Further validation is needed to fully understand the underlying mechanism, such as eCLIP-seq of different RNA binding proteins and profiling of RNA degradome. Thirdly, we found the m6A levels increase after meiosis resumption in the oocytes, however, the reason and biological functions of the relatively low m6A level in the fully-grown oocytes remain unknown. It is now known that DNA methylation is erased from the primordial germ cells (PGCs) during their transition to primordial oocytes, and the de novo DNA methylation takes place during the follicle growth62, but the regulation role played by m6A during oocyte growth and early embryo development remains unknown. It will be very interesting to explore this issue by profiling the m6A of PGCs and oocytes obtained from follicles at different developmental stages.
ZGA is an early hallmark of early embryonic development and may be controlled by the fertilization time, spatial location, cell cycle counts, and cell size63. The timing of ZGA differ among different species. In mice, minor ZGA occurs from the late 1-cell stage, and major ZGA occurs at the late 2-cell stage, and moreover, the minor ZGA is required for a normal major ZGA process20,64. Although the molecular bias of lncRNAs25,53, histone modification65, and molecules from the second polar body66 have been studied in the 2-cell embryos, the difference of ZGA process among the individual blastomeres has not been extensively studied. Our m6A landscape of single blastomeres at 2-cell stage by scm6A-seq revealed the asynchronous ZGA process in the 2-cell embryos. This outcome can be partially explained by the different cell cycle progression between the two blastomeres in a 2-cell embryo. In addition, this is not surprising because of the appearance of many 3-cell embryos during mouse embryonic development54,67. The differences between the two blastomeres is a maternal trait induced by the asymmetric distribution of m6A methylome. As m6A modified RNA tend to form ribonucleoprotein (RNP) complex by m6A readers68,69. While RNPs have an important role in RNA metabolism and gene expression regulation in embryos and embryonic stem cells70,71, we speculate that there is an asymmetric distribution of m6A-related RNPs in the blastomeres in the 2-cell embryos. On the other hand, the details of how m6A methylome regulates the ZGA process and cell fate decision need further clarification.
Overall, scm6A-seq provides a robust tool to explore the functions of the RNA m6A modification during embryo development at the single-cell level. Similar strategies might also be expanded to other RNA modifications with reliable antibodies.
Methods
Animals maintenance
The construction of Mettl3 conditional knockout mice in oocytes (Mettl3flox/flox;Gdf9-cre, referred to as Mettl3 cKO) referenced from the previous report15. The Mettl3flox/flox female mice were used as the control group (referred to as control). All mice described above were kept at C57BL/6J genetic background and housed under specific pathogen-free (SPF) conditions with 12 light/12 dark cycles at 22 °C, 40% humidity. All animal experiments were approved by the Animal Care and Use Committee of China Agricultural University.
Collection and culture of oocytes
The control and Mettl3Gdf9 cKO oocytes and zygotes are collected as descripted before. Briefly, 4- to 6-week-old females of both control and Mettl3Gdf9 cKO mice were injected with 5 IU pregnant mare serum gonadotropin (PMSG) (Solarbio, P9970). For GV oocytes, the cumulus cell-oocyte complex was collected and the oocytes (>70 μm) were released with microcapillary pipettes in M2 media 44 to 48 h after PMSG injection. For MII oocytes, the mice were then injected with 5 IU of human chorionic gonadotropin (hCG) (Ningbo No.2 hormone factory, Zhejiang, China). Then the MII oocytes were collected 14–16 h later. For zygotes, the mice were mated with male mice with known fertility, and successful mating was confirmed by the presence of vaginal plugs. For Mettl3Gdf9 cKO zygotes, the sample is further confirmed by the morphology of the pronuclei.
For in vitro maturation, the collected GV oocytes from 6- to 8-week-old wild-type mice were cultured in MEM medium (Gibco, 11095072) with slight modifications72. The GVBD oocytes and MI oocytes were collected after 4 and 8 h, respectively. The MII oocytes were collected after 12 h when the fist polar body appears.
Embryo collection and single blastomere isolation
Embryos were collected from 4- to 6-week-old C5LNB6 females mated with C5LNB6 males. To induce ovulation, females were administered 5 IU of hCG intraperitoneally, 44–48 h post injection of 5 IU of PMSG. Embryos were collected from female mice at the following time points post hCG injection: one-cell stage (19–21 h post hCG), early two-cell stage (31–32 h post hCG), mid-two-cell stage (39–40 h post hCG), late two-cell stage (46–48 h post hCG), and four-cell stage (54–56 h post hCG). To obtain the single cell of embryos, the zona pellucida was removed using Tyrode’s solution and then the embryos were treated with 0.25% Trypsin-EDTA for several seconds. The single cell was then put in the lysis buffer for scm6A-seq.
Immunofluorescence staining
Oocytes were fixed in 4% PFA for 30 min at room temperature (RT) followed by permeabilization in 0.25% Triton X-100 for 15 min before blocking in 3% BSA for 1 h at RT. Then the oocytes were incubated with primary antibodies (Anti METTL3, Abcam, Cat # ab195352, 1:500 in washing solution) in 3% BSA overnight at 4 °C. The embryos were washed 3 times for 5 min each in washing solution (0.1% Tween-20 in PBS) and incubated with donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 conjugated (Invitrogen, Cat # A21206, 1:500 in washing solution) for 1 h at RT. After washed thoroughly for three times in washing solution, embryos were stained with DAPI (1:1000 in washing solution) for 5 min and subjected to take images using a confocal microscope.
Library preparation for scm6A-seq
All adenylated adapters were synthesized using a 5’ DNA Adenylation Kit (NEB, M2611A) according to the manufacturer’s instructions, and stored at −20 °C for no more than six months. Single cells were piped into 96-well plates with 5 µl lysis buffer (0.5 µl of 10× lysis buffer (TaKaRa, 635013), 2.5 µl 2×FPE Buffer (Vazyme, N402), 0.5 µl of 40 U/ml RNase inhibitor, murine (NEB, M0314L), 0.5 µl of gDNA eraser from HiFiScript gDNA Removal cDNA Synthesis Kit (CWBIO, CW2582M), and 1 µl of nuclease-free water. The samples were kept at RT for 3 min for cell lysis before gDNA was removed by gDNA eraser at 42 °C for 5 min. Then, RNA was fragmented to approximately 200 nt by heating at 94 °C for 8 min. The gDNA eraser was deactivated by heating at 75 °C. Then, T4 PNK and PNK buffer (NEB, M0201 L) were added to each reaction at 37 °C for 45 min to remove the RNA 3’ phosphate group before RNA adapter ligation. For the ligation reaction, 2 µl T4 ligation buffer, 1 µl T4 RNA ligase 2, truncated K227Q (NEB, M0373L), 7 µl 50% PEG8000 (NEB, M0373L), 1 µl adenylated adapter (with barcode 1), and 1 µl RNase inhibitor (NEB, M0314L) were added to each tube. RNA ligation was performed at 4 °C overnight with gently shaking at 350 rpm. Then, RNA from all wells was pooled together and purified using 1.5× RNA Clean XP beads (Beckman, A63987) after inactivating the T4 RNA ligase by heating at 75 °C for 20 min. For the second-round labeling, the well-ligated samples were pooled together and purified with 1× RNA clean beads. The mixed samples are annealed with adapters with barcode 2 before gap filling with T4 DNA polymerase at 12 °C for 15 min according to the instructions (NEB, M0203L). The RNAs from different samples were pooled together and divided into two parts: one-fifth were retained for Input (designated as RNA-seq), the remaining four-fifths were subjected to m6A enrichment through incubating with m6A antibody and protein A-G beads according to the instructions of Magna MeRIP m6A Kit (Merck, 17-10499). After m6A immunoprecipitation (IP), the m6A-containing RNAs were eluted from the beads with N6-Methyladenosine-5’-monophosphate sodium salt (designated as m6A-IP) and the supernatant were also collected and sequenced as the background for m6A detection. The RNAs from input, supernatant, and m6A-IP samples were extracted, dissolved in 40 µl nuclease-free water, and subjected to reverse transcription and the template-switching reaction using picoRT oligo and 5’ blocked TSO. The cDNAs were purified using 1× DNA clean beads and eluted in 32 µl nuclease-free water. Then, 40 µl 2× KAPA HiFi HotStart ReadyMix (KAPA, KK2602), 4 µl pico-RT primer, and 4 µl T7 primer were added for the first PCR amplification. Then, the T7-promoter-inserted libraries were transcribed into RNAs with T7 polymerase and further purified using RNA clean beads (Vazyme, N412-01). Then, rRNA products were removed using an RNase H-based method as described previously73. Then, the rRNA-minus RNAs were annealed and reverse transcribed using pico-RT primer. Finally, the cDNA was amplified using sequencing adapters (NEB, E7335L) before library purification with 1× DNA clean beads (Vazyme, N411-01). Library sequencing was performed on the Illumina NovaSeq6000 system. Notably, for the 10 individual GV oocytes as data quality control, the m6A-containing RNAs were extracted by proteinase K digestion instead of competitive elution after m6A-IP. All the oligos used were ordered from GeneScript. The oligo sequences were provided in Supplementary Data 9.
Library construction for bulk MeRIP-seq and RNA-seq
For bulk MeRIP-seq and RNA-seq of 25, 50, and 70 GVs, the RNA was extracted using RNeasy Plus Kits (Qiagen, 74034) according to the instruction ignoring the DNA filter step. Then the total RNA was subjected to m6A enrichment according to procedures of scm6A-seq except for without cell labeling. After m6A immunoprecipitation, the m6A-Protein A/G beads-RNA complexes were washed first by low salt buffer followed by high salt buffer and m6A binding buffer. Then the m6A-modified RNA fragments were extracted by proteinase K digestion and acid phenol-chloroform as described previously15. The library construction was also performed as for scm6A-seq. Sequencing was performed on an Illumina NovaSeq 6000 platform.
scm6A-seq data preprocessing
The template switch oligo (TSO) sequence of raw sequencing reads was trimmed by Cutadapt74 (version 2.10) with parameters ‘-g TCCGATCT -O8 -n5 -e 0 --action trim’, ‘-g CACGTCTC’ for multiple TSO sequences filtering, and multiple adapter sequences trimming with parameters ‘-a NNNNNNNAGATCGGA -a TCGGAAGAGCACAC -A NNNNNNNAGATCGGA -A TCGGAAGAGCACAC -e0 -j 6 -m 28 -O8’. Then reads of individual cells were separated by barcode sequences (Supplementary Data 10) using a homemade script. First, the reads starting with “[barcode]NNNNGGG” were selected, and other reads were eliminated. For single sample data, the four-base random sequence and the first seven bases at read 2 were combined and wrote at the first line of the fastq-format and defined as unique molecular identifiers (UMIs). To remove the PCR duplicate reads, the first seven bases at the beginning of read 1 and read 2 were combined together as a molecular barcode. Then, the molecular barcodes were sorted, and the unique reads were retained. For single-cell parallel sequencing data, the index barcode, barcodes at read 1 and read 2 were decoded step by step. The data are separated into individual files, corresponding to each single cell. All scripts used in this study are available from the corresponding author by request.
After single-cell data separation, the reads of m6A-IP, supernatant, and RNA-seq from scm6A-seq and bulk sequencing were aligned to the Mus musculus (mouse) genome (GRCm38/mm10) from Ensembl75 (release 96) using HISAT276 (version 2.0.5). Samtools (version 1.6)77 was used to filter and sort the mapped reads, then featureCounts (version 2.0.3)78 program was applied to summarize the gene counts in each sample, and fragments per kilobase per million mapped reads (FPKM), transcripts per kilobase of exon model per million mapped reads (TPM) and counts of exon model per million mapped reads (CPM) genes expression were calculated by StringTie (version 2.2.1)79. The relative m6A level of each cell is defined as log2(reads count (m6A-IP/(m6A-IP + RNA-seq + Supernatant))).
Saturation analysis for scm6A-seq data
Sequencing saturation of scm6A-seq was evaluated by random re-sampling approach. The aligned bam files of all reads in each GV oocyte were annotated to mm10 genome by BEDTools (version 2.30.0)80. Then, shuf of Linux was used to randomly select a given number of reads from total reads and the number of mapped reads covered genes was counted. The given number of reads and corresponding covered genes were applied to build saturation curve for scm6A-seq.
Metagene analysis for scm6A-seq data
The relative position of each transcription start site (TSS) and stop codon in the corresponding transcript were identified in Ensembl mm10 genome (release 96), respectively. For metagene density plots, bigwig files were converted from bam files using bamCoverage of deepTools81 (version 3.3.1) with 5 bp bins, then computeMatrix commond was used to calculate scores per genome region with the parameter ‘reference-point --referencePoint TSS’, and ‘-b 500 -a 500 -R ${StopCondon}’ for the stop codon or ‘-b 50 -a 500 -R ${TSS}’ for TSS sites in each transcript. Finally, the ratio of m6A-IP to RNA-seq from scm6A-seq in the score matrix files was calculated and visualized by density plot. To generate the metagene profiles for each stage, Samtools was used to merge single-cell data from scm6A-seq.
Identification of high-confident m6A peaks
MACS2 callpeak82 (version 2.1.4) was used for m6A-enriched peaks calling for m6A-IP with corresponding RNA-seq as control in scm6A-seq and bulk sequencing. The parameters of ‘--keep-dup all --nomodel -g mm -B -q 0.05’ were used for peak calling. Furthermore, high-confident m6A peaks were identified when peaks were annotated to transcriptome of Ensembl mm10 (release 96) with ‘fold_enrichment > 2 && −log10(q value) > 10’ in the peak calling files.
For scm6A-seq, only the peaks identified in both RNA-seq and supernatant as control samples were considered high-confident and used for further analysis by intersectBed of BEDTools (version 2.26.0) with ‘-f 0.51 -F 0.51’. Merged data was used as input samples for identification of m6A peaks in each stage. METTL3-dependent m6A peaks in GV oocytes were defined as enriched peaks only identified in control compared with the Mettl3Gdf9 cKO samples with expression more than 1 (FPKM ≥ 1).
For bulk15 and low input32 m6A sequencing data, the parameters of ‘--keep-dup all --nomodel -g mm -B -q 0.05’ were used for peak calling. The m6A peaks were then annotated to transcriptome of Ensembl mm10 (release 96) for the identification of the m6A-modified RNAs in each replicate.
m6A motif enrichment analysis
Motif discovery for filtered m6A peaks was identified by findMotifsGenome.pl of HOMER software (version 4.11.1). The sequences extracted inside the m6A peaks from the transcriptome was used as target sequences and the randomly shuffling peaks upon whole transcriptome were obtained by shuffleBed of BEDTools.
Differential expression of METTL3-dependent m6A modified RNAs
Differentially expressed genes between Mettl3Gdf9 cKO and control oocytes were calculated by edgeR83 with the P value <0.05. For further confirm, Integrative Genomics Viewer (IGV)84 was used to display the m6A signals and RNA abundance of scm6A-seq in mm10 genome with bigwig files generated by BEDTools.
Clustering analysis of scm6A-seq data
Before clustering analysis, gene expression from single-cell and merged samples of scm6A-seq among different oocytes and embryo developmental stages was z score-normalized. Clustering of gene sets during oocyte maturation and early embryonic development was performed by K-means algorithm85 with 10,000 iterations. Hierarchical clustering analysis was applied to classify cell samples using hclust function in R program. The principal component analysis (PCA) and uwot of uniform manifold approximation and projection (UMAP) method86 was used for dimensionality reduction and cell clusters of single-cell data. For further clustering of the scm6A-seq data by comprehensive analysis of the gene expression level and RNA m6A level, the single cell matrix after dimensionality reduction by UMAP using monocle287 and the log2((IP reads)/(Total Reads)) values were used for hierarchical clustering.
m6A reader analysis
YTHDF2/3 binding peaks identified by eCLIP-seq in mESCs were downloaded from GEO database (GSE151788)42. IGF2BP2/3 binding peaks of hESCs were extracted from eCLIP-seq data (GSE78509)52. UCSC liftOver88 was used to convert genome coordinates between human and mouse. shuffleBed of BEDTools was applied to randomly permute the genomic locations of m6A reader binding peaks. Distance between m6A readers’ targets and identified m6A peaks was calculated using the “closet” command of BEDTools.
Translatome and proteome analysis
The translation signals and efficiency of different developmental stages for each gene were calculated from public data (GSE169632)89. Translation signals were defined as gene expression levels calculated from LiRibo-seq data. Translation efficiency (TE) of m6A-modified RNAs was evaluated by ratio of RPKM between LiRibo-seq and RNA-seq samples in different developmental stages. The protein abundance of oocyte maturation, mESCs, and early embryonic developmental stages were obtained from LC-MS/MS51 and TMT-based quantitative MS assay data90, respectively.
Maternal and ZGA genes analysis
Genes expressed in MII with FPKM ≥ 1 were considered as maternal genes. Maternal decay genes were identified by comparing RNA-seq in scm6A-seq of MII and zygote using threshold of log2(fold change) > 0.5 and P.adj < 0.05 in DESeq291 with count tables from featureCounts. Minor ZGA genes were identified as genes upregulated in early and mid-2-cell stages with the previous adjacent developmental time point as control (log2(fold change) > 0.5 and P.adj < 0.05). Major ZGA genes were defined by comparing the late 2-cell and 4-cell embryos with log2(fold change) > 0.5 and P.adj < 0.05 to samples of previous adjacent developmental time point, respectively. K-means algorithm was also used for clustering of maternal and ZGA genes during early embryonic development.
Functional enrichment analysis
For the functional annotation of gene sets, DAVID92, Matascape (version 3.5)93, and ClueGO94 of cytoscape95 were applied to annotate Gene Ontology Consortium96 biological processes with threshold P value <0.05 without special instructions.
Transcriptome factors analysis
Transcription factors and their corresponding targets in mice were collected from TRRUST database97 (version 2), and their interaction network was visualized by Cytoscape (version 3.9.1). UpsetR (version 1.4.0)98 was applied to display the overlapped RNAs among different developmental stages.
To analyze the TFs responsible for ZGA. The differently expressed genes are further filtered with threshold log2(FPKM(Class2)/FPKM(Class1)) > 2. Then the sequence inside ATAC-seq peaks in the promoter region and gene body of these genes are used for TFs enrichment using MEME-Chip (version 5.5). The ATAC-seq peaks are identified using public data (GSE66581) of the 2-cell embryos using MACS2 with default parameters.
Statistics and reproducibility
All statistical analyses were performed using R program and Prism software. Statistically significant differences between different groups were evaluated by one- or two-sided test (Wilcoxon test, Hypergeometric test, or Student t test) without additional adjustments. All significant levels are presented in figures as singe asterisk (*) when P value < 0.05, **P < 0.01, and ***P < 0.001, unless otherwise specified. More than 10 cells of each stage of oocytes/embryos are subjected to scm6A-seq. And more than 4 oocytes of each stage were collected for single-cell RNA-seq.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data generated in this study have been deposited in Genome Sequence Archive of National Genomics Data Center under accession code CRA006425. The expression value, m6A peaks, and other necessary matrix data generated in this study are provided in the Supplementary Information and Source Data files. The immunofluorescence staining (IF) data generated in this study have been deposited in Mendeley (https://data.mendeley.com/datasets/dhskwr3r5f/draft?a=69360c62-93e5-415a-b5cf-36b51dcfe0f9). Source data are provided with this paper.
Code availability
The main code and demo test data have been deposited in Zenodo (https://zenodo.org/record/6809317#.YsfA0IRByUk). Other codes are available from the lead contact upon request.
References
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6, 28 (2017).
Liu, J. et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020).
Liu, J. D. et al. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 591, 322–326 (2021).
Wei, J. B. et al. FTO mediates LINE1 m6A demethylation and chromatin regulation in mESCs and mouse development. Science 376, 968–973 (2022).
Kim, J. & Lee, G. A. Metabolic control of m6A RNA modification. Metabolites 11, 11 (2021).
Hu, L. L. et al. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 40, 1210–1219 (2022).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Meyer, K. D. DART-seq: an antibody-free method for global m6A detection. Nat. Methods 16, 1275–1280 (2019).
Zeng, Y. et al. Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 16, e2006092 (2018).
Zhang, Z. et al. Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 5, eaax0250 (2019).
Jiang, X. L. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 6, 16 (2021).
Zhao, B. X. S. et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017).
Mu, H. Y. et al. METTL3-mediated mRNA N6-methyladenosine is required for oocyte and follicle development in mice. Cell Death Dis. 12, 989 (2021).
Ivanova, I. et al. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67, 1059–1067 (2017).
Hu, Y. et al. Oocyte competence is maintained by m6A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell Death Differ. 27, 2468–2483 (2020).
Sui, X. S. et al. METTL3-mediated m6A is required for murine oocyte maturation and maternal-to-zygotic transition. Cell Cycle 19, 391–404 (2020).
Du, Z., Zhang, K. & Xie, W. Epigenetic reprogramming in early animal development. Cold Spring Harb. Perspect. Biol. 14, a039677 (2022).
Liu, B. F. et al. The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 587, 139–144 (2020).
Xia, W. K. & Xie, W. Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Rep. 15, 1158–1175 (2020).
Xu, R. M., Li, C., Liu, X. Y. & Gao, S. R. Insights into epigenetic patterns in mammalian early embryos. Protein Cell 12, 7–28 (2021).
Sha, Q. Q. et al. Characterization of zygotic genome activation-dependent maternal mRNA clearance in mouse. Nucleic Acids Res. 48, 879–894 (2020).
Zhao, L. W. et al. Nuclear poly(A) binding protein 1 (PABPN1) mediates zygotic genome activation-dependent maternal mRNA clearance during mouse early embryonic development. Nucleic Acids Res. 50, 458–472 (2022).
Wang, J. Q. et al. Asymmetric expression of LincGET biases cell fate in two-cell mouse embryos. Cell 175, 1887–1901 (2018).
White, M. D., Zenker, J., Bissiere, S. & Plachta, N. Instructions for assembling the early mammalian embryo. Developmental Cell 45, 667–679 (2018).
Deng, Q., Ramskold, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013).
Yang, Q. et al. Single-cell CAS-seq reveals a class of short PIWI-interacting RNAs in human oocytes. Nat. Commun. 10, 3389 (2019).
Xiong, Z. et al. Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat. Cell Biol. 24, 968–980 (2022).
Zou, Z. et al. Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science 378, abo7923 (2022).
Wu, Y. et al. N6-methyladenosine regulates maternal RNA maintenance in oocytes and timely RNA decay during mouse maternal-to-zygotic transition. Nat. Cell Biol. 24, 917–927 (2022).
Dierks, D. et al. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat. Methods 18, 1060–1067 (2021).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–U284 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Moshitch-Moshkovitz, S., Dominissini, D. & Rechavi, G. The epitranscriptome toolbox. Cell 185, 764–776 (2022).
Yao, H., Yang, Y. & Yang, Y. G. scDART-seq: mapping m6A at the single-cell level. Mol. Cell 82, 713–715 (2022).
Tegowski, M., Flamand, M. N. & Meyer, K. D. scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 82, 868–878 (2022).
Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Lasman, L. et al. Context-dependent functional compensation between Ythdf m6A reader proteins. Genes Dev. 34, 1373–1391 (2020).
Ma, J. Y. et al. Maternal factors required for oocyte developmental competence in mice: transcriptome analysis of non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) oocytes. Cell Cycle 12, 1928–1938 (2013).
Sun, M. J. et al. An essential role for the intra-oocyte MAPK activity in the NSN-to-SN transition of germinal vesicle chromatin configuration in porcine oocytes. Sci. Rep. 6, 13 (2016).
Xu, Q. H. et al. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat. Genet. 51, 844–856 (2019).
Zhang, B. J. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016).
Dai, X. X. et al. A combinatorial code for mRNA 3’UTR-mediated translational control in the mouse oocyte. Nucleic Acids Res. 47, 328–340 (2019).
Fan, H. Y. & Sun, Q. Y. in The Ovary 181–203 (2019).
Li, A. et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447 (2017).
Shi, H. L. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Wang, S. F. et al. Proteome of mouse oocytes at different developmental stages. Proc. Natl Acad. Sci. USA 107, 17639–17644 (2010).
Conway, A. E. et al. Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep. 15, 666–679 (2016).
Hupalowska, A. et al. CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell 175, 1902–1916 (2018).
Rossant, J., Chazaud, C. & Yamanaka, Y. Lineage allocation and asymmetries in the early mouse embryo. Philos. Trans. R. Soc. B-Biol. Sci. 358, 1341–1349 (2003).
Kim, K., Park, S. & Roh, S. Lipid-rich blastomeres in the two-cell stage of porcine parthenotes show bias toward contributing to the embryonic part. Anim. Reprod. Sci. 130, 91–98 (2012).
Park, S. K., Won, C., Choi, Y. J., Kang, H. & Roh, S. The leading blastomere of the 2-cell stage parthenogenetic porcine embryo contributes to the abembryonic part first. J. Vet. Med. Sci. 71, 569–576 (2009).
Wu, J. Y. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016).
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Takada, Y., Iyyappan, R., Susor, A. & Kotani, T. Posttranscriptional regulation of maternal Pou5f1/Oct4 during mouse oogenesis and early embryogenesis. Histochemistry Cell Biol. 154, 609–620 (2020).
Lee, J., Hyeon, D. Y. & Hwang, D. Single-cell multiomics: technologies and data analysis methods. Exp. Mol. Med. 52, 1428–1442 (2020).
Mendel, M. et al. Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol. Cell 71, 986–1000 e1011 (2018).
Seisenberger, S. et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 (2012).
Schulz, K. N. & Harrison, M. M. Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234 (2019).
Abe, K. et al. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl Acad. Sci. USA 115, E6780–E6788 (2018).
Torres-Padilla, M. E., Parfitt, D. E., Kouzarides, T. & Zernicka-Goetz, M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218 (2007).
Jin, H. B. et al. The second polar body contributes to the fate asymmetry in the mouse embryo. Natl Sci. Rev. 9, 11 (2022).
Cicconet, M., Gutwein, M., Gunsalus, K. C. & Geiger, D. Label free cell-tracking and division detection based on 2D time-lapse images for lineage analysis of early embryo development. Computers Biol. Med. 51, 24–34 (2014).
Edupuganti, R. R. et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017).
Fu, Y. & Zhuang, X. W. m6A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 16, 955–963 (2020).
Tian, Q., Wang, X. F., Xie, S. M., Yin, Y. & Zhou, L. Q. H3.3 impedes zygotic transcriptional program activated by Dux. Biochem. Biophys. Res. Commun. 522, 422–427 (2020).
Zhang, G. Q. et al. Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat. Commun. 13, 16 (2022).
Guo, J. et al. Oocyte stage-specific effects of MTOR determine granulosa cell fate and oocyte quality in mice. Proc. Natl Acad. Sci. USA 115, E5326–E5333 (2018).
Levin, J. Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–U767 (2010).
Marcel, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Yates, A. D. et al. Ensembl 2020. Nucleic Acids Res. 48, D682–D688 (2020).
Kim, D., Landmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–U121 (2015).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Feng, J. X., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinforma. 14, 178–192 (2013).
Demidenko, E. The next-generation K-means algorithm. Stat. Anal. Data Min. 11, 153–166 (2018).
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J Open Source Softw. 3, 861 (2018).
Qiu, X. J. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017).
Kuhn, R. M., Haussler, D. & Kent, W. J. The UCSC genome browser and associated tools. Brief. Bioinforma. 14, 144–161 (2013).
Zhang, C. X., Wang, M., Li, Y. S. & Zhang, Y. Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition. Sci. Adv. 8, 14 (2022).
Gao, Y. W. et al. Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep. 21, 3957–3969 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 38 (2014).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Zhou, Y. Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Han, H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386 (2018).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Acknowledgements
This work was supported by grants from the Innovative Research Group Project of the National Natural Science Foundation of China (32121001 Y.G.Y.), the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA16010501 Y.G.Y., XDB38020500 Y. Y., XDPB2004 Y.Y.), the CAS Project for Young Scientists in Basic Research (YSBR-073 Y.G.Y.), National Key R&D Program of China (2019YFA0110900 Y.P.S., 2019YFA0110901 Y.G.Y., 2019YFA0802201 Y.S.C.), and the National Natural Science Foundation of China (92053115 Y.Y., 91940304 Y.Y.), CAS Key Research Projects of the Frontier Science (QYZDY-SSW-SMC027 Y.G.Y.), the Youth Innovation Promotion Association of CAS (2018133 Y.Y.), the Beijing Nova Program (Z201100006820104 Y.Y., 20220484210 Y.Y.), and the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01 Y.G.Y.), and the K. C. Wong Education Foundation (Y.G.Y.).
Author information
Authors and Affiliations
Contributions
H.Y. performed the experiments with the help of J.W.X., G.G.S., Y.J.G., and D.B.L.; D.R.Z. and Q.L. collected the oocytes and embryos. Y.T.C. helped to check the chromatin configuration of GV oocytes; C.C.G. performed bioinformatics analysis with critical input from X.F. and Y.S.C.; Y.G.Y., Y.P.S., Y.Y., and J.Y.H. conceived this project, supervised the study and interpreted the data. Y.G.Y., Y.Y., C.C.G., Y.L.Z., L.H., H.Y., wrote the manuscript, and with contributions from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yao, H., Gao, CC., Zhang, D. et al. scm6A-seq reveals single-cell landscapes of the dynamic m6A during oocyte maturation and early embryonic development. Nat Commun 14, 315 (2023). https://doi.org/10.1038/s41467-023-35958-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-35958-7
This article is cited by
-
Decoding epitranscriptomic regulation of viral infection: mapping of RNA N6-methyladenosine by advanced sequencing technologies
Cellular & Molecular Biology Letters (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.