Abstract
FANCM suppresses crossovers in plants by unwinding recombination intermediates. In wheat, crossovers are skewed toward the chromosome ends, thus limiting generation of novel allelic combinations. Here, we observe that FANCM maintains the obligate crossover in tetraploid and hexaploid wheat, thus ensuring that every chromosome pair exhibits at least one crossover, by localizing class I crossover protein HEI10 at pachytene. FANCM also suppresses class II crossovers that increased 2.6-fold in fancm msh5 quadruple mutants. These data are consistent with a role for FANCM in second-end capture of class I designated crossover sites, whilst FANCM is also required to promote formation of non-crossovers. In hexaploid wheat, genetic mapping reveals that crossovers increase by 31% in fancm compared to wild type, indicating that fancm could be an effective tool to accelerate breeding. Crossover rate differences in fancm correlate with wild type crossover distributions, suggesting that chromatin may influence the recombination landscape in similar ways in both wild type and fancm.
Similar content being viewed by others
Introduction
Meiotic recombination is initiated by numerous programmed DNA double-strand breaks (DSBs), catalyzed by SPO11 and MTOPVIB1,2, that are repaired by homologous recombination into crossovers (COs) or non-crossovers (NCOs). COs are characterized by the reciprocal exchange of DNA between homologous chromosomes, whereas NCOs occur at sites of non-reciprocal DNA repair using either the homologue or the sister-chromatid as a template3. In Arabidopsis and wheat, COs form via the class I or class II pathways4,5,6,7. The class I pathway accounts for ~85% COs and is required to maintain CO assurance, thus formation of the obligate CO. The class I pathway ensures that each chromosome pair receives at least one CO through the action of meiosis specific proteins MER3/RCK, MSH4, MSH5, SHOC1, ZIP4, PTD and HEI106, thereby promoting correct chromosome segregation. Class I COs are interference-sensitive and therefore more likely to be spaced further apart than would be expected by random chance8,9,10. Class II COs are interference-insensitive, accounting for ~15% COs and are partially dependent on the MUS81 endonuclease11,12.
The number of COs per chromosome is tightly constrained in the majority of eukaryotes (~1–3), regardless of physical chromosome size5,13. DSBs usually occur far in excess of COs in plants, suggesting that they do not limit CO formation. In hexaploid wheat the skewed ratio of ~2100 DSBs to ~42 COs per cell reveals that only 2% of potential repair sites mature into COs14, indicating that underlying mechanisms negatively regulate CO formation. In Arabidopsis, class I COs are limited by expression of HEI1015, the presence of PPX116, and the installation of the synaptonemal complex transverse filament proteins ZYP1a/ZYP1b17,18. Class II COs are limited by three independent, anti-recombination pathways involving FANCM, FIDGETIN and RECQ419,20,21,22. The FANCM (Fanconi anemia complementation group M homolog) helicase functions with MHF1 and MHF2 to limit COs by unwinding inter-homolog repair intermediates, such as D-loops, and promotes repair as NCOs, in inbred lines19,20,21,23,24. Ablation of FANCM in Arabidopsis restores bivalent formation in class I CO deficient mutants to near wild-type levels19,20. Single fancm mutants exhibited a 3-fold increase in COs by pollen fluorescent marker analysis19, but chiasmata (the cytological manifestations of COs) were significantly reduced20, although technical limitations may have precluded detection of more than one closely spaced CO using this analysis. A significant reduction in the number of bivalents and loss of the obligate chiasma was recently observed in lettuce fancm mutants25, although Brassica (diploid and allotetraploid), rice (Oryza sativa) and pea (Pisum sativum) fancm mutants displayed increased COs26,27.
Wheat is an allopolyploid crop species where the predominantly distally distributed COs/chiasmata rarely exceed two per chromosome pair7,14,28,29. COs mainly form in gene-rich regions30 that contain a high frequency of TIR-Mariner transposons31 and lower levels of DNA polymorphism32. COs also correlate with enrichment of the Polycomb histone modification H3K27me3 at the distal ends, indicating a potential role for facultative heterochromatin in shaping the recombination landscape33. CO frequency and distribution reduce the probability of creating advantageous novel allelic combinations in crop breeding programmes. Therefore, anti-CO factors, such as FANCM provide a potential way to overcome these limitations34. Here, using wheat fancm null mutants and VIGS, we demonstrate that class I COs are reduced in number, resulting in loss of the obligate CO, but this is offset by an increase in class II COs.
Results
FANCM is conserved in wheat
To identify FANCM orthologues in wheat, BLAST searches were performed using the Arabidopsis thaliana FANCM amino acid sequence. Two FANCM orthologues were identified in tetraploid wheat T. turgidum: TtFANCM-A1 (TRITD4Av1G171480) and TtFANCM-B1 (TRITD4Bv1G035000). Three FANCM orthologues were identified in hexaploid wheat T. aestivum: TaFANCM-A1 (TraesCS4A02G217700), TaFANCM-B1 (TraesCS4B02G096400) and TaFANCM-D1 (TraesCS4D02G092800). All copies are full-length and predicted to produce functional proteins. FANCM is highly conserved between ploidy levels with FANCM-A1 and FANCMB-1 primary amino acid sequences being identical in tetraploid and hexaploid wheat (1447/1447 & 1458/1458, respectively), while the three FANCM homoeologues, FANCM-A1, FANCMB-1 and FANCM-D1, share 94.2% amino acid identity, with polymorphisms at 85 residues (1377/1462). A consensus sequence was created from the three wheat homoeologues to compare with Arabidopsis. Wheat FANCM shares 38.8% (543/1502) overall amino acid identity with Arabidopsis, with increased homology in the predicted DEXDc (DEAD-like helicases superfamily; 70.7%; 135/191) and HELICc (helicase superfamily c-terminal; 67.9%; 93/137) domains.
FANCM is required for normal fertility
Single fancm homoeolog mutants for tetraploid (Triticum turgidum, AABB) (Fig. 1a) and hexaploid wheat (Triticum aestivum, AABBDD) (Fig. 1b) were crossed to create null knockouts. The tetraploid Ttfancm_1 null exhibited a 36% reduction in total number of seeds per plant from 66 ± 2 (n = 4) in wild type to 43 ± 5 (n = 4) in the mutant (p < 0.05, Mann–Whitney U Test; Supplementary Fig. 1). The hexaploid Tafancm null exhibited a 15% reduction in seeds per plant from 164 ± 4 (n = 6) in wild type to 139 ± 6 (n = 6) in the mutant (p < 0.05, Mann–Whitney U Test; Supplementary Fig. 1). Pollen viability decreased from 92% in wild type (n = 2042) to 76% in Ttfancm_1 (n = 1994) (Supplementary Fig. 2). This indicates that defects occurring during gamete formation had a direct impact on seed development.
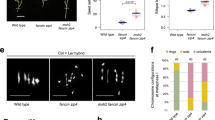
a TtFANCM coding region for the A and B homoelogs with conserved domains shown (DEAD-like helicases superfamily (DEXDc) and helicase superfamily c-terminal (HELICc)), and TILLING mutations (red) indicated. b TaFANCM coding region for the A, B and D homoelogs with conserved domains shown (DEXDc and HELICc), and TILLING mutations (red) and VIGS target sites (black) indicated. c Box plot of chiasmata frequency per male meiocyte. Box extends from the 25th to 75th percentile, the centre line is the median value and the whiskers denote min and max values. The number of meiocytes sampled for each line is shown in brackets. n.s. = p > 0.05. *** = p < 0.001 (Pairwise Wilcoxon Rank Sum Test with Bonferroni correction). d Proportion of rings, rods and univalents per male meiocyte. The number of meiocytes sampled for each line is shown in brackets. Source data are provided as a Source Data file.
FANCM is required for crossover assurance
To determine the cause for the reduction in fertility and pollen viability in fancm null mutants, a cytological analysis was performed on tetraploid wheat meiocytes. At meiotic metaphase I (MI) the number of chiasmata, rod and ring bivalents was indistinguishable between wild type and the single homoeolog mutants (Ttfancm-A1_m1, Ttfancm-A1_m2, and Ttfancm-B1; p values > 0.05, Pairwise Wilcoxon Rank Sum Tests). However, in Ttfancm_1 nulls, chromosome pairs without chiasmata at MI revealed loss of the obligate chiasma and defective CO assurance (Fig. 1c,d, Supplementary Fig. 3, Supplementary Data 3). In Ttfancm_1, 0.98 ± 0.09 (n = 104) pairs of univalents per cell were observed at MI, compared to only 0.05 ± 0.03 (n = 60) in wild type (p < 0.001, Pairwise Wilcoxon Rank Sum Test), and 64% of cells possessed at least one univalent pair in Ttfancm_1, compared with only 5% in wild type. The number of rod bivalents per cell also increased from 1.7 ± 0.16 (n = 60) in wild type to 4.7 ± 0.09 (n = 104) in Ttfancm_1 (p < 0.001, Pairwise Wilcoxon Rank Sum Test). The increase in univalent pairs and rod bivalents coincided with an 18% decrease in chiasmata per cell, from 26 ± 0.2 (n = 60) in wild type to 22 ± 0.3 (n = 104) in Ttfancm_1 (p < 0.001, Pairwise Wilcoxon Rank Sum Test). There was no significant difference between Ttfancm_1 and Ttfancm_2 null mutants at MI (p values > 0.05, Pairwise Wilcoxon Rank Sum Tests). Chromosome pairs without chiasmata mis-segregated in Ttfancm_1, resulting in unbalanced gametes (Supplementary Fig. 4). At anaphase I, chromosome mis-segregation was observed in 43% of cells in Ttfancm_1 (n = 40), compared with 2.5% in wild type (n = 40). Chromosome mis-segregation through loss of the obligate chiasma is the most likely cause for reduced pollen viability and seed production in fancm null mutants.
A comparative cytological analysis was then performed on fancm null mutants in hexaploid wheat. In Tafancm, 0.53 ± 0.09 (n = 81) univalents per cell were observed, compared to 0.01 ± 0.01 (n = 76) in wild type (p < 0.001 Pairwise Wilcoxon Rank Sum Test), and 37% of cells possessed at least one univalent pair, compared with 1.3% in wild type (Fig. 1c,d, Supplementary Fig. 3, Supplementary Data 3). The number of rod bivalents per cell also increased from 1.9 ± 0.16 (n = 76) in wild type to 4.6 ± 0.26 (n = 81) in Tafancm (p < 0.001, Mann–Whitney U Test). The increase in frequency of univalents and rod bivalents coincided with a 9% decrease in total chiasmata per cell, from 40 ± 0.2 (n = 76) in wild type to 36 ± 0.4 (n = 81) in Tafancm (p < 0.001, Mann–Whitney U Test), indicating that the requirement for FANCM in maintaining CO assurance is similar in both tetraploid and hexaploid wheat.
As the TILLING mutants have a high density of background mutations that may affect the role of FANCM based on previous studies investigating heterozygosity21,24, TaFANCM-Virus Induced Gene Silencing (VIGS) was performed on hexaploid wheat. Plants inoculated with the Barley Stripe Mosaic Virus:TaFANCM-i construct exhibited 0.58 ± 0.14 (n = 33) univalents per cell, compared to 0.02 ± 0.02 (n = 50) in the empty virus control (p < 0.001, Pairwise Wilcoxon Rank Sum Test), and 40% of cells possessed at least one univalent pair, compared with 2% in the control (Fig. 1c,d, Supplementary Fig. 3, Supplementary Data 3). The number of rod bivalents also increased from 1.56 ± 0.18 (n = 50) in the control to 4.24 ± 0.33 (n = 33) in BSMV:TaFANCM-i (p < 0.001, Pairwise Wilcoxon Rank Sum Test). The increase in frequency of univalents and rod bivalents coincided with a 9% decrease in total chiasmata per cell, from 42 ± 0.2 (n = 50) in the control to 38 ± 0.5 (n = 33) in BSMV:TaFANCM-i (p < 0.001, Pairwise Wilcoxon Rank Sum Test). The same effect was observed in a second, independent construct (BSMV:TaFANCM-ii) where there was no significant difference with construct (i) at MI (p values > 0.05, Pairwise Wilcoxon Rank Sum Tests), indicating that the VIGS and TILLING mutants reproduced the same phenotype for chiasma formation.
The distribution of remaining chiasmata in fancm null mutants, while still deviating significantly from a Poisson distribution (χ2(26) = 38.9, n = 104, p < 0.05; Fig. 3b), spanned a greater range than wild type (23–28 WT vs 16–27 Ttfancm_1), thus implying a defect in CO control. Therefore, an analysis utilizing pSc119.2-2, 5 S rDNA and 45 S rDNA synthetic oligonucleotide probes was performed on Ttfancm_2 MI chromosomes to determine if there was a pattern in the reduction of chiasmata. Chromosomes 1B and 6B possess 45 S rDNA sites that form the Nucleolar Organization Regions (NOR) and can therefore be reliably identified7,35 (Supplementary Fig. 5). Chromosomes 1B and 6B exhibited a significant reduction in the number of chiasmata per chromosome in Ttfancm_2 compared to wild type ((1.7 ± 0.07, n = 45 to 1.3 ± 0.11, n = 40 on 1B (p < 0.01, Mann–Whitney U Test) and 1.8 ± 0.06, n = 45 to 1.3 ± 0.11, n = 40 on 6B (p < 0.001, Mann–Whitney U Test; Supplementary Fig. 5)). The Fluorescence in situ hybridization (FISH) analysis also revealed a chromosomal bias in chiasmata distribution in the wild type, as chromosomes 1B and 6B were overrepresented among rod bivalents. They accounted for 27%, indicating that chromosomes with NORs were more likely to form a single chiasma than other chromosomes (χ2(2) = 13.16, p < 0.01). Chromosomes 1B and 6B were also overrepresented among the univalent pairs in the Ttfancm_2 null mutant (χ2(2) = 21.07, p < 0.01), accounting for 44% of all observed. However, the overall reduction in chiasmata in Ttfancm_2 occurred throughout all chromosomes and 1B and 6B did not significantly deviate from expected values (χ2(2) = 5.83, p > 0.05). Therefore, a direct consequence of the fancm null mutant appears to be a reduced ability of the NOR chromosomes to buffer against a general loss of chiasmata.
FANCM is required for localization of HEI10 at late prophase I
As CO assurance was abrogated in the fancm null mutants, localization dynamics of the class I CO protein HEI10 were investigated. HEI10 initially localizes as small, numerous, axis-associated foci during leptotene in Arabidopsis and wheat, that coarsen into fewer, larger foci, that mark class I CO sites at late prophase I29,36,37. At leptotene, the number of HEI10 foci per meiocyte was not significantly different between Ttfancm_1 (356 ± 32, n = 7) and wild type (351 ± 25, n = 7) (p > 0.05, Mann–Whitney U Test) (Supplementary Fig. 6). At zygotene, the number of HEI10 foci was not significantly different between Ttfancm_1 (156 ± 14, n = 7) and wild type (156 ± 13, n = 9) (p > 0.05, Mann–Whitney U Test) (Supplementary Fig. 6). However, at pachytene HEI10 foci decreased by 20% from 39 ± 0.8 (n = 82) in wild type to 32 ± 0.5 (n = 74) in Ttfancm_1 (Mann–Whitney U Test, p < 0.001), (Fig. 2). Furthermore, at diakinesis HEI10 foci reduced by 29%, from 34 ± 0.5 (n = 54) in wild type to 24 ± 0.5 (n = 60) in Ttfancm_1 (Mann–Whitney U Test, p < 0.001) (Fig. 2) and this also occurred in hexaploid wheat. At pachytene the number of HEI10 foci per meiocyte decreased by 9%, from 58 ± 0.6 (n = 54) in wild type to 53 ± 1.2 (n = 34) in the Tafancm null (p < 0.001, Mann–Whitney U Test; Fig. 2). This reduction in HEI10 foci at pachytene suggests that formation of a proportion of class I COs is sensitive to the loss of FANCM in wheat.
a–f Co-immunofluorescence of HEI10 (white) and ZYP1 (red) on meiotic prophase I chromosome spreads. Representative micrographs are shown from replicates, (a) (n = 82), (b) (n = 74), (c) (n = 54), (d) (n = 60), (e) (n = 54), and (f) (n = 34). Pachytene (a, c, e and f) and early diakinesis (b and d). Scale bars = 10 µm. g Counts of HEI10 foci per cell with mean values ± SD. The number of meiocytes sampled for each line is shown in brackets. *** = p < 0.001 (Mann–Whitney U Test). Source data are provided as a Source Data file.
DSBs, meiotic progression, and SC formation appear normal in fancm
As a proxy marker for DSB formation during meiosis, localization of the strand-exchange protein RAD5138 was scored during leptotene. The number of RAD51 foci was not significantly different between Ttfancm_1 (1397 ± 21, n = 5) and wild type (1403 ± 24, n = 5) (p > 0.05, Mann–Whitney U Test) (Supplementary Fig. 7), indicating no apparent effect on the number of early recombination sites. MSH5, an early class I CO recombination protein7,39 localized to unsynapsed and synapsed chromosomes at zygotene, with no significant difference in the number of foci between Ttfancm_1 (336 ± 14 n = 5) and wild type (378 ± 22, n = 5) (p > 0.05, Mann–Whitney U Test) (Supplementary Fig. 8).
Fixed anthers of equivalent sizes from wild type and Ttfancm_1 corresponded to the same meiotic stages, indicating that there was no delay in meiotic progression. In both cases, anthers 0.6–0.7 mm in length contained cells at leptotene, 0.8 mm at zygotene, 0.9 mm at pachytene, 0.95–1.1 mm at MI, 1.1 mm at dyad, and 1.3 mm at tetrad. Furthermore, immunolocalisation of ASYNAPSIS1 (ASY1)40 and ZYP141 revealed normal axis formation and synapsis in Ttfancm_1, as previously described for wild type7,29,42. In wild type and Ttfancm_1, ASY1 formed a linear signal on the unsynapsed chromosome axes at leptotene, which depleted during zygotene on synapsed chromosomes (Supplementary Fig. 9). ZYP1 was initially detected during late-leptotene as presynaptic foci that extended throughout zygotene until a complete linear signal was observed along synapsed chromosomes at pachytene. These data indicate that early stages of meiotic recombination are unperturbed in the fancm null mutants.
FANCM limits class II crossovers
TtMSH5B possesses a natural loss-of-function deletion mutation and therefore the single Ttmsh5a mutant produces a null allele of MutS\({{{{{\rm{\gamma }}}}}}\) that is defective in class I COs7. The Ttmsh5 null was crossed with the Ttfancm_1 null to create the quadruple null Ttmsh5 Ttfancm to investigate whether fancm mutants can compensate for loss of class I COs. In Ttmsh5, 4.29 ± 0.13 (n = 194) bivalents per cell were observed, which increased ~2.2-fold to 9.34 ± 0.24 (n = 86) in Ttmsh5 Ttfancm (p < 0.001, Pairwise Wilcoxon Rank Sum Test). Furthermore, ring bivalents accounted for only 3.2% of chromosome pairs in Ttmsh5, which significantly increased to 21% in Ttmsh5 Ttfancm (p < 0.001, Pairwise Wilcoxon Rank Sum Test). The increase in ring bivalents coincided with a ~2.6-fold increase in the number of chiasmata per cell, from 4.7 ± 0.15 (n = 194) in Ttmsh5 to 12.4 ± 0.41 (n = 86) in Ttmsh5 Ttfancm (p < 0.001, Pairwise Wilcoxon Rank Sum Test; Fig. 1c,d, Supplementary Fig. 3, Supplementary Data 3). The additional chiasmata in Ttmsh5 Ttfancm did not deviate significantly from a Poisson-predicted distribution (χ2(20) = 9.74, n = 86, p > 0.05; Fig. 3d), implying that they formed via the interference-insensitive class II CO pathway.
a–d Observed and Poisson-predicted distributions of chiasma frequency per cell. a The observed wild-type distribution of chiasmata deviates significantly from a Poisson-predicted distribution (χ2(28) = 113.16, n = 60, p < 0.01, Chi-squared test). b The distribution of chiasmata in Ttfancm_1 deviates significantly from a Poisson-predicted distribution (χ2(26) = 38.9, n = 104, p < 0.05, Chi-squared test). c The distribution of chiasmata in Ttmsh5 does not deviate significantly from a Poisson-predicted distribution (χ2(14) = 5.63, n = 194, p ≥ 0.975, Chi-squared test). d The distribution of chiasmata in Ttmsh5 Ttfancm_1 does not deviate significantly from a Poisson-predicted distribution (χ2(20) = 9.74, n = 86, p > 0.95, Chi-squared test). Source data are provided as a Source Data file.
FANCM localizes to meiotic chromosomes during prophase I
To gain further insight into the role of FANCM during meiotic recombination, a wheat FANCM antibody was generated and its specificity verified by immunolocalisation. FANCM was first detected at late-leptotene as numerous (991 ± 42, n = 5), small (0.4 µm ± 0.02, n = 50), axis-associated foci (Fig. 4a). By early-zygotene fewer (20 ± 1.3, n = 22), but larger (0.7 µm ± 0.02, n = 50) distinct FANCM foci were observed of which 58% (254/441) associated with ASY1 and 42% (187/441) with ZYP1 (Fig. 4b), indicating localization to synapsed and unsynapsed chromosomes. By mid-zygotene, the number of large FANCM foci peaked at 29 ± 1.5 (n = 29), which is similar to the number of designated class I CO sites7 (Fig. 4c). FANCM foci then decreased to 18 ± 1.8 (n = 11) at late-zygotene (Fig. 4d), and then 12 ± 1.0 (n = 12) by pachytene (Fig. 4e). When co-immunostained with anti-TaASY1, anti-TaFANCM also gave a linear signal to unsynapsed chromosomes, which was not present when used exclusively or in combination with other antibodies (e.g., anti-HvHEI10, anti-AtZYP1), indicating that cross-reactivity with ASY1 may be responsible for the linear signal. Furthermore, the linear signal was present in the Ttfancm_1 null mutant, whereas the FANCM foci were absent (Fig. 4f).
a–g Co-immunofluorescence of FANCM (red), ASY1 (green) and ZYP1 (blue) on meiotic prophase I chromosome spreads. h Co-immunofluorescence of FANCM (red), HEI10 (green) and ZYP1 (blue) on meiotic chromosome spreads at mid-zygotene. Representative micrographs are shown from replicates, (a) (n = 5), (b) (n = 22), (c) (n = 29), (d) (n = 11), (e) (n = 12), (f) (n = 16), (g) (n = 20), (h) (n = 5). Insets, 2× magnified view of outlined region, showing co-localization of FANCM and HEI10 foci on synapsed chromosomes. Scale bars = 10 µm.
As the number of zygotene FANCM foci correlated with the number of designated class I CO sites, FANCM was co-immunostained with HEI10. Due to difficulties staging the meiocytes without ASY1, an alternative anti-HvHEI10 was used which produces a linear signal on unsynapsed chromosomes and forms prominent foci on synapsed chromosomes7. As the HEI10 foci on unsynapsed chromosomes were obscured by the non-specific linear signal, only FANCM foci localizing on synapsed chromosomes were scored. A mean of 14 ± 2.7 (n = 5) FANCM foci per cell were observed of which 8 ± 2.2 (59%) overlapped with emergent HEI10 foci (Fig. 4h). Furthermore, FANCM immunolocalisation to class I-deficient mutant Ttmsh5 displayed a 51% reduction in the number of FANCM foci at mid-zygotene, from 29 ± 1.5 (n = 29) in wild type to 15 ± 0.8 (n = 20) in Ttmsh5 (Fig. 4g). Taken together, these data are consistent with 51–59% FANCM foci being present at designated class I CO sites and promoting class I interfering COs, while 41–49% FANCM foci are located at potential NCO sites.
Genetic mapping reveals an overall increase in crossovers in fancm mutants
Genetic mapping using molecular marker analysis was performed on segregating tetraploid and hexaploid fancm null mutants as an additional tool to measure recombination frequency and distribution. The hexaploid wheat Cadenza fancm null mutant was crossed with Avalon to produce F2 plants and the F3 segregants were used for genetic map construction (Fig. 5a). Segregating Cadenza/Avalon genetic markers on the 35 K Axiom chip provided 21 intervals on ten chromosomes that were tested for recombination using a LOD 8 threshold. This revealed an increase in COs (>10%) in 11 intervals, a reduction in five (>−10%) and no difference in 4 (between 10% and −10%) (Table 1, Fig. 5b, Supplementary Data 4, 5 & 6). Chromosome 1 A exhibited the largest increase in genetic map length in the fancm null mutant and is shown as an example (Fig. 5c). The map length was increased by 19.1% in fancm over WT when adding the cM distances across all regions, or by 31.3% when averaging the percentage cM increase from the 21 intervals.
a Crossing schematic for the development of the triple homozygous mutant and F3 mapping populations fixed at fancm for either wild type or mutant alleles. b Segregating regions in both F3 populations aligned and ordered by RefSeqv1.0 coordinates (Mbp), flanking markers indicated by lines. Increasing regions (>10%) are highlighted with shades of orange and decreasing regions (<−10%) in blue, with neutral regions (−10 to +10%) in white. The location of fancm on chromosome 4 A is indicated by a red line and centromeric regions are shaded in gray. c Comparison of the genetic maps of the F3 populations for chromosome 1 A (cM). The control population is aligned to the physical positions (Mbp) to illustrate chromosomal location.
Recombination in tetraploid wheat fancm mutants was measured with KASP markers designed against mapped segregating SNPs from the mutation donor lines. Four F3 populations, two homozygous WT at FANCM and two fixed homozygous for the mutations, consisting of 95 plants, were tested across seven genetic intervals on 5 chromosomes. Recombination was increased in the chromosome 1B interval but decreased within the 2 A, 2B and 3 A intervals, but were the same in another 3 A interval and 3B (Table 2, Supplementary Data 7). The trend was an overall decrease, but statistically the mutant and wild type were indistinguishable in genetic map length for the intervals tested.
Crossover rate differences in fancm correlate wild type crossover distributions
Genomic regional variation in CO rate differences between fancm and wild type shows a significant positive correlation with a previously published wild type CO rate map derived from a Chinese Spring × Renan recombinant inbred population43 (Supplementary Fig. 10). Differential CO rate in fancm also shows non-significant positive relationships with ChIP-seq signals for the chromosome axis protein ASY1 and the meiotic recombinase DMC1, the euchromatic marks H3K4me1, H3K4me3 and H3K27ac, and the Polycomb mark H3K27me3, and non-significant negative correlations with H3K9me2, H3K27me1, centromeric CENH3, and DNA methylation in CG and CHG contexts (Supplementary Fig. 10, Supplementary Data 8 and 9). Therefore, while COs are increased in fancm, their genomic distribution is comparable to that in wild type. This suggests that chromatin may influence the recombination landscape in similar ways in both wild type and fancm.
Discussion
In the absence of FANCM, 64% of metaphase I cells in the tetraploid and 37% in the hexaploid possessed at least one univalent pair, thus indicating loss of CO assurance. This led to chromosome mis-segregation during meiosis II, followed by a decrease in pollen viability and seed production. The metaphase I FISH analysis revealed an overall reduction in chiasmata, suggesting a random loss of COs, consistent with studies in lettuce fancm mutants25. Nucleolar Organizing Region containing chromosomes 1B and 6B exhibited a lower level of chiasmata in wild type and exhibited univalents more frequently in fancm than other chromosomes. The loss of the obligate chiasma and reduction of HEI10 foci from 34 to 24 during diakinesis in tetraploid wheat suggested that loss of FANCM had a direct effect on the class CO I pathway. These data are consistent with the increase of rod bivalents observed in A. thaliana fancm mutants20, and increased numbers of univalents observed in lettuce fancm25.
FANCM localized as small, numerous foci during leptotene that coarsened into fewer, large foci during zygotene that correlated with the number of expected COs. This pattern is consistent with FANCM localizing to influence the fate of a proportion of recombination intermediates. FANCM could be required to release resected DSB ends from invasion of sister chromatids to enable engagement with the homolog, as reported in budding yeast44. However, reduced inter-homolog engagement of the first strand may not have a significant effect on CO formation in wheat, due to the ~50:1 excess of DSBs over COs14, which may mask this effect, compared to budding yeast where the ratio of DSBs to COs is ~1.7:145. This effect could be tested by crossing wheat fancm and spo11 hypomorphic mutants to systematically reduce the number of DSBs, thus enabling comparison of inter-homolog engagement in spo11 and fancm/spo11 mutants. If there was an early role for FANCM a greater loss of inter-homolog engagement would be expected in the fancm/spo11 mutants compared to spo11.
In tetraploid wheat, ~29 FANCM foci coarsened during zygotene of which ~51–59% localized with HEI10. In the absence of FANCM, HEI10 foci reduced from 34 to 24 per nucleus at diakinesis, suggesting that FANCM is required to promote 29% of class I COs. Our data are consistent with a model in which FANCM facilitates second-end capture at class I CO sites. FANCM may release the DSB second-end from association with the sister-chromatid, or other homologous/homoeologous templates, similarly to budding yeast, but at a later stage. This model is consistent with current data from rice where FANCM interacts with RPA1a during meiosis to promote CO formation46, and in Arabidopsis RPA1a plays a crucial role in second-end capture47. Our data supports the molecular function of FANCM as a DNA helicase that unwinds strand invasion D-loop intermediates during zygotene. The requirement for FANCM to function at a subset of designated class I CO sites may be due to a stochastic propensity for DSB second-ends to invade the sister chromatid, rather than the homolog. This would also imply that CO site designation may occur at the D-loop stage in wheat.
A role for FANCM in limiting the class II CO pathway was revealed by crossing the mutant with the class I CO mutant msh57. Chiasmata increased over ~2.6-fold in the fancm/msh5 quadruple mutant compared to the msh5 mutant, although this was insufficient to restore CO levels, as reported in Arabidopsis19,20. Therefore, FANCM may unwind a small proportion of D-loop intermediates between homologous chromosomes that were destined to form NCOs, but in its absence may be processed by the class II CO pathway.
These data suggest that FANCM reduces the probability of non-interfering class II CO’s from forming, whilst promoting interfering class I CO’s, thus producing a landscape where COs are spaced further apart. FANCM appears to ensure the fate of a subset of recombination intermediates, rather than being involved in CO interference per se, but a recent analysis of the role of FANCM and its interacting partner FANCD2 in Arabidopsis suggests that it directly promotes interference of class I COs, based on HEI10 foci frequency and distribution25. However, due to the significant reduction of HEI10 foci in wheat, this effect may be obscured, but still active.
The data presented here suggest that FANCM is required at CO designated sites/NCOs during zygotene. In wild type tetraploid wheat, the ~10-fold excess of HEI10 foci at mid-zygotene (351) compared to the eventual (34) foci at diakinesis superficially should be sufficient to enable CO assurance. However, in the absence of the 29 FANCM foci (of which ~51–59% localized with HEI10), the eventual HEI10 foci reduced to 24 at diakinesis in the mutant. Therefore, these data are consistent with class I CO designation occurring before FANCM coarsens at mid-zygotene, as the majority of the remaining HEI10 were unable to compensate. Recent studies have shown that CO interference is mediated by ZYP1 in Arabidopsis17,18, although modelling of CO designation with the coarsening of HEI10 foci occurring during mid/late prophase I may also play a role37. The FANCM data here indicates that CO designation occurs earlier than mid-zygotene, consistent with budding yeast3. This also raises the question of how FANCM is regulated to function at both designated class I CO sites and non-designated sites to ensure the optimum fate of recombination intermediates.
The wheat fancm mutants exhibited a reduction in fertility (36% in tetraploid and 15% in hexaploid wheat), due to loss of the obligate chiasma. This is consistent with a previous study utilizing FANCM-VIGS on tetraploid wheat F1 hybrids that showed a significant reduction in fertility, but no differences in recombination on chromosome 1A48. The immunohistochemistry, cytological and molecular marker analysis presented here reveal that loss of class I COs are offset by an increase of class II COs in the tetraploid, thus resulting in no net change. However, fewer HEI10 foci were reduced in hexaploid wheat fancm mutants, compared to the tetraploid, leading to an overall increase of COs by 31%. Based on an increase of class II chiasmata detected in the tetraploid, it is likely that the increase in COs in the hexaploid arose via the class II CO pathway. The molecular marker data revealed that COs increased in the majority of intervals tested, thus providing an opportunity to modulate recombination in wheat breeding programs. The distal H3K27me3-enriched regions experienced the greatest increase in COs, whereas the interstitial/proximal regions were more likely to be lower, similar to recombination increases in the barley recq4 mutant49.
Methods
Identification of wheat FANCM
Wheat FANCM orthologues were identified using the Arabidopsis thaliana amino acid sequences (At1g35530) to BLAST against publicly available databases: Triticum turgidum50 Svevo.v1 https://plants.ensembl.org/Triticum_turgidum) and T. aestivum43 IWGSC https://plants.ensembl.org/Triticum_aestivum). Wheat cds were aligned using the Clustal W algorithm (gap open cost = 12, gap extend cost = 3), translated and the primary sequence ran through the Conserved Domain Database (NCBI).
Plant material
Triticum turgidum ‘Kronos’ was used as a wild-type control for experiments involving the tetraploid Kronos TILLING mutant lines obtained from www.SeedStor.ac.uk51. We selected knockout mutants (premature termination codon; Supplementary Data 1) in TtFANCM-A1 (TRITD4Av1G171480) exon 2 (Kronos3873; abbreviated K3873 hereafter) and exon 8 (K0234). Similarly, we selected a knockout mutant (K3842) in the third exon of TtFANCM-B1 (TRITD4Bv1G035000). We confirmed the mutations via genotyping (Supplementary Data 2) and named the single mutants as Ttfancm-A1_m1 (K3873), Ttfancm-A1_m2 (K0234), and Ttfancm-B1 (K3842). We also generated double null mutants: Ttfancm_1 (K3873 x K3842) and Ttfancm_2 (K0234 x K3842) and characterized a Ttmsh5-A1 mutant (K863), which is functionally a null (Ttmsh5) as there is a natural loss-of-function deletion in TtMSH5-B1 (Desjardins et al. 2020), and a Ttfancm Ttmsh5 double mutant (K3873 × K3842 × K863). Triticum aestivum cv. ‘Cadenza’ was used as a wild-type control for experiments involving hexaploid Cadenza TILLING mutant lines. We selected knockout mutants in all three TaFANCM homoeologs including TaFANCM-A1 (TraesCS4A02G217700; Cadenza0641, abbreviated C0641 hereafter), TaFANCM-B1 (TraesCS4B02G096400; C1195) and TaFANCM-D1 (TraesCS4D02G092800; C0827). The single mutants were crossed to generate a Tafancm null mutant (C0641 × C1195 × C0827)52. Plants were grown under controlled environmental growth conditions: photoperiod 16 h, temperature 21 °C (day)/16 °C (night) and relative humidity ~60%. VIGS experiments were conducted in a Level 3 biological containment facility at Rothamsted Research.
RT-PCR
Total RNA was extracted from T. turgidum cv. ‘Kronos’ and T. aestivum cv. ‘Cadenza’ spikes using ISOLATE II RNA Mini Kit (Bioline), and cDNA was synthesized with the Tetro cDNA synthesis kit (Bioline). The coding sequences for both varieties were amplified and sequenced using homoeolog-specific primers: FANCM_A_F 5′-CTGGATGTTGGCTGCACTCG-3′ and FANCM_A_R 5′- ATGTGGTTGCTTTCAGAGGTA-3′, FANCM_B_F 5′-AGAGGCTATGTTTCTATCACC-3′ and FANCM_B_R 5′-GATCCTGATGTATTCCCTACC-3′, FANCM_D_F 5′-AAGGAAGGAAGTGGGAAAGT-3′ and FANCM_D_R 5′- CCAGGCAAGCATGAATATCC-3′.
Virus induced gene silencing
The barley stripe mosaic virus, virus induced gene silencing system (BSMV:VIGS)53,54 was used to target expression of TaFANCM in T. aestivum ‘Bobwhite’. Two non-overlapping regions with high predicted silencing efficiency were selected, following an in silico analysis of TaFANCM-A1 cds (TraesCS4A02G217700) with si-Fi21 (siRNA Finder). For the first selected region, primers TaFANCM-vigs-i_F 5′-AAGGAAGTTTAAGTGGACTCATTCACCAGGAG-3′ and TaFANCM-vigs-i_R 5′-AACCACCACCACCGTTCATCGCCACGGACAGCA-3′ were used to amplify 259–572 bp of TraesCS4A02G217700 coding region with Q5 DNA polymerase (NEB). For the second region, primers TaFANCM-vigs-ii_F 5′-AAGGAAGTTTAAGTGAAGGACCAGAGCTGCAAG-3′ and TaFANCM-vigs-ii_R 5′-AACCACCACCACCGTGCCTGAACTGAAGTACTGAGC-3′ were used to amplify 2102–2376 bp of TraesCS4D02G092800 coding region with Q5 DNA polymerase (NEB). The amplicons were cloned into pCa-γbLIC to create a recombinant BSMV RNAγ. The recombinant pCa-γbLIC, as well pCaBS-α and pCaBS-β, were then transformed into electrocompetent Agrobacterium tumefaciens (GV3101), agroinfiltrated, and the virus accumulated in intermediate host Nicotiana benthamiana, prior to inoculation of wheat plants with infected sap at the 4–4.5 leaf stage (~28 days post-sowing)”54. For cytological analysis, anthers were dissected and fixed in 3:1 (v/v) ethanol: acetic acid at 14-days post inoculation.
FANCM antibody production
Primers AbF 5′-AGCATATGTTGGTGGCAGCTGGTGTG-3′ and AbR 5′-TCCTCGAGATTGTATTCCCCAGCTCG-3′ were used to amplify 1107–1935 bp of the TraesCS4B02G096400 (FANCM-B1) coding region with Q5 DNA polymerase (NEB). The PCR product was ligated into pDrive (Qiagen) and sequenced (Eurofins). pDrive was then digested with NdeI and XhoI, and the insert was ligated into pET21b (Merck-Millipore). Transformed BL21 (DE3) cells expressed a 32 kDa recombinant protein that was used to produce a rat anti-FANCM antibody (DC Biosciences).
Fertility assessment
Fertility was assessed in Ttfancm_1 and Tafancm null mutants by counting the total number of seeds per plant and comparing with wild type using Mann–Whitney U tests (Minitab v 18.1.0.0). The ratio of viable to non-viable pollen grains was also determined for Ttfancm_1 using Alexander staining55.
Cytological procedures
Meiotic chromosome spreads were prepared from fixed anthers. Following digestion (30–60 min at 37 °C) in citrate buffer containing 0.33% cellulase (Duchefa Biochemie) and 0.33% pectolyase (MP Biomedicals), the anthers were macerated in a drop of water, spread in 60–80% acetic acid at 45 °C, and refixed in 3:1 (v/v) ethanol: acetic acid54,56. Slides were mounted in VECTASHIELD® Mounting Medium with DAPI (Vector Labaratories). Fluorescence in situ hybridization (FISH) was conducted using three labelled synthetic oligonucleotides: pSc119.2-2 [A488]35, pTa794-1 (5 S) [TxRd]7 and pTa71-1 (45 S) [A647]35. Nikon Ni-E and Eclipse Ci fluorescence microscopes equipped with NIS elements software were used to capture and quantify images. Chiasmata number was interpreted according to bivalent shape29. Rod bivalents were scored as a minimum of 1 and ring bivalents scored as a minimum of 2, with additional chiasmata scored based on shape.
Chromosome spreads for HEI10 immunostaining were prepared as above. For all other antibodies, chromosome spreads were prepared from fresh anthers. Following digestion (8 min at 37 °C) in enzyme mixture containing 0.4% cytohelicase from Helix pomatia (Sigma-Aldrich), 1.5% sucrose and 1% polyvinylpolypyrrolidone (Sigma-Aldrich), meiocytes were squeezed out of anthers using a brass rod, spread in 0.4–1% lipsol detergent (Appleton Woods Ltd) and fixed in 4% ice-cold paraformaldehyde (Fisher Scientific)54. Immunostaining was conducted using the following primary antibodies: anti-TaASY1 guinea pig, 1:500;7 anti-AtZYP1C rat, 1:500;41 anti-AtZYP1C rabbit, 1:500;57 anti-AtRAD51 rabbit, 1:200;58 anti-MSH5 rabbit12, 1:200;7 anti-HvHEI10 rabbit, 1:2507. Secondary antibodies that were used, 1:200: goat anti-guinea pig AMCA (Jackson ImmunoResearch; 106-155-003); goat anti-guinea pig Alexa Fluor® 488 (Abcam; #A-11073); goat anti-guinea pig Alexa Fluor® 594 (Abcam; #A-11076); goat anti-rat AMCA (Jackson ImmunoResearch; 112-155-003); goat anti-rat Alexa Fluor® 594 (Invitrogen; #A-11007); goat anti-rabbit AMCA (Jackson ImmunoResearch; 111-155-003); goat anti-rabbit Alexa Fluor® 488 (Invitrogen; #A-11008) and goat anti-rabbit DyLight® 594 (Vector Labs; DI-1595). Meiocytes were staged with anti-ZYP1 or anti-ASY1 to ensure focal counts were made at equivalent stages; for anti-RAD51 (leptotene), anti-MSH5 (zygotene), and anti-HEI10 (leptotene to diakinesis). Counts were performed using NIS software and significance established using Pairwise Wilcoxon Rank Sum with Bonferroni correction (RStudio v 1.2.5033), Mann–Whitney U (Minitab v 18.1.0.0) or Chi-squared (χ2) tests, as appropriate.
Kronos tetraploid F3 marker-based recombination analysis
Fancm double null mutants (Ttfancm_1) were developed by crossing plants with premature stop mutations in the A (Kronos3873.chr4A.517401432) and B (Kronos3842.chr4B.100316098) genomes (Supplementary Data 1). F1 plants were checked for heterozygosity with mutation-specific KASP markers (Supplementary Data 2) and self-pollinated for F2 seed. Four F2 sibling plants, two fixed for WT FANCM and two fixed for mutations in the A&B homoeologs (Ttfancm_1) were self-pollinated to produce F3 seed. Genome-wide mutations from donor TILLING lines were extracted (http://www.wheat-tilling.com/51, with regions and primers selected for screening based on chromosome position and genome specificity (Supplementary Data 2). KASP primers were tested on the F2 plants to check for heterozygosity in fourteen selected regions, seven of which were found to be segregating in at least one WT and one fancm double mutant population. F3 populations of 95 plants were sown for the four F2 lines (two fixed for WT FANCM and two fixed for the A and B homoeolog fancm mutations). The seven regions were screened for recombination analysis with either two or three markers per region. After genotyping, the final number of lines for analysis was determined by excluding missing lines or lines with missing data, and the recombination distance calculated (recombinants/total progeny × 2) × 100.
Avalon x Cadenza hexaploid F3 marker-based recombination analysis
Fancm triple mutants (Tafancm) were produced by crossing mutants carrying premature stop mutations from the three homoeologs (Cadenza0641.chr4A.517403720, Cadenza1195.chr4B.100321720 and Cadenza0827.chr4D.67793583) and tracking mutations with mutation-specific KASP markers. Triple mutant Tafancm homozygous plants were crossed with Avalon and the F1 progeny self-pollinated. F2 siblings fixed as either homozygous wild type for the three FANCM homoeologs, or homozygous for all three mutations at fancm were self-pollinated to obtain F3 seed. F3 populations of 90 and 91 plants respectively, were sown from populations arising from fixed WT FANCM and triple mutant fancm F2 siblings. F3 seed from an Avalon × Cadenza cross were germinated and grown in pots filled with peat-based soil and kept in a glasshouse at 15–18 °C with 16 h light, 8 h dark. Leaf-tissue was harvested from individual plants 14 days post-sowing, when the plants were at an early seedling stage. DNA was extracted following the protocol59 with minor modifications. DNA concentration was assessed using a Qubit 2.0 Fluorometer and was then normalized to 23 ng/µl ready for analysis with the Axiom® Wheat Breeder’s array59. Sample preparation for array genotyping was performed with the Beckman Coulter Biomek FX. Samples were then genotyped using the Axiom® 35 K Wheat Breeders array in conjunction with the GeneTitan® using standard Affymetrix protocols59. Axiom Analysis Suite (version 3.1.51.0) was used to assign genotype calls. Axiom 35 K genotyping data was ordered by chromosome and physical position obtained from IWGSC RefSeq v.1.0 (IWGSC, 2018). Monomorphic markers (>90% of plants carrying either the Cadenza or Avalon allele), markers with high levels of missing data (>20%), markers with distorted segregation (heterozygous < 25% or >75%) and dominant markers (Allele 0/2 < 5) were removed prior to mapping. The remaining segregating markers were compared for both WT and triple mutant fancm populations. A total of 13 regions were selected to investigate recombination frequency based on the presence of identical markers segregating in both populations. Genetic mapping was carried out for each region individually using MSTmap online (http://www.mstmap.org/) using a LOD 8 threshold. Where linkage groups contained identical markers, cM distance was compared between populations. In some cases, selected regions were split into two or more linkage groups. The ‘cM map length increase’ was calculated as the percentage increase of the total fancm cM distance over WT control across all regions and the ‘cumulative percentage increase’ describes the average of the percentage difference of all chromosome regions.
Associating FANCM crossovers with chromatin marks
Differential CO rate (fancm cM/Mb minus wild type cM/Mb), wild type CO rate43, and chromatin and meiotic protein mean signals, profiled via ChIP-seq or bisulfite-seq (as analyzed in ref. 33), were calculated within each genetic marker interval. Spearman’s rank-order correlation coefficients (rs) were computed for each parameter pair and plotted as a correlation matrix, with rs denoted by cell color (R v 4.0.0). P values for rs correlation coefficients were standardized to represent those based on pairwise values across 100 marker intervals and are indicated within each cell of the correlation matrix. Included data sets are differential CO rate (fancm cM/Mb minus wild type cM/Mb; “Diff_cMMb”), wild type CO rate derived from a Chinese Spring x Renan genetic map (“IWGSC_cMMb”)43, ASY1, DMC1, H3K4me3, H3K9me2 and H3K27me1 ChIP-seq33, H3K4me1 and H3K27ac ChIP-seq60, H3K27me3 and H3K36me3 ChIP-seq43, CENH3 ChIP-seq61, whole-genome bisulfite sequencing-derived DNA methylation (mCG, mCHH and mCHG proportions)43, and the distance between the midpoint of each marker interval and the midpoint of previously defined centromeric coordinates (“Dist_to_CEN”)43. Histone ChIP-seq and bisulfite-seq data are derived from Chinese Spring seedlings or leaf tissue61,43,60,33, and ASY1 and DMC1 ChIP-seq data are derived from Chinese Spring immature pre-emergence spikes33.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data supporting the findings of this work are available within the paper and its Supplementary Information files. A reporting summary for this paper is available as a Supplementary Information file. Raw data of the images are available from the corresponding author upon request. Source data are provided with the paper. Source data are provided with this paper.
References
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997).
Vrielynck, N. et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351, 939–943 (2016).
Borner, G. V., Kleckner, N. & Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004).
Osman, K., Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J. & Franklin, F. C. Pathways to meiotic recombination in Arabidopsis thaliana. N. Phytologist 190, 523–544 (2011).
Mercier, R., Mezard, C., Jenczewski, E., Macaisne, N. & Grelon, M. The molecular biology of meiosis in plants. Annu Rev. Plant Biol. 66, 297–327 (2015).
Wang, Y. & Copenhaver, G. P. Meiotic Recombination: Mixing It Up in Plants. Annu Rev. Plant Biol. 69, 577–609 (2018).
Desjardins, S. D. et al. MutS homologue 4andMutS homologue 5Maintain the Obligate Crossover in Wheat Despite Stepwise Gene Loss following Polyploidization(1)([CC-BY]). Plant Physiology 183, 1545–1558 (2020).
Higgins, J. D., Armstrong, S. J., Franklin, F. C. & Jones, G. H. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570 (2004).
Jones, G. H. & Franklin, F. C. Meiotic crossing-over: obligation and interference. Cell 126, 246–248 (2006).
Berchowitz, L. E. & Copenhaver, G. P. Genetic interference: don’t stand so close to me. Curr. Genom. 11, 91–102 (2010).
Berchowitz, L. E., Francis, K. E., Bey, A. L. & Copenhaver, G. P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. Plos Genet. 3, e132 (2007).
Higgins, J. D., Buckling, E. F., Franklin, F. C. & Jones, G. H. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162 (2008).
Fernandes, J. B., Seguela-Arnaud, M., Larcheveque, C., Lloyd, A. H. & Mercier, R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl Acad. Sci. USA 115, 2431–2436 (2018).
Gardiner, L. J. et al. Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biol. 20, 69 (2019).
Ziolkowski, P. A. et al. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 31, 306–317 (2017).
Nageswaran, D. C. et al. HIGH CROSSOVER RATE1 encodes PROTEIN PHOSPHATASE X1 and restricts meiotic crossovers in Arabidopsis. Nat. Plants 7, 452–467 (2021).
Capilla-Perez, L. et al. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl Acad. Sci. USA 118, https://doi.org/10.1073/pnas.2023613118 (2021).
France, M. G. et al. ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis. Proc. Natl Acad. Sci USA 118, https://doi.org/10.1073/pnas.2021671118 (2021).
Crismani, W. et al. FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012).
Knoll, A. et al. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24, 1448–1464 (2012).
Girard, C. et al. AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms. Plos Genet. 11, e1005369 (2015).
Seguela-Arnaud, M. et al. Multiple mechanisms limit meiotic crossovers: TOP3alpha and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl Acad. Sci. USA 112, 4713–4718 (2015).
Girard, C. et al. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 42, 9087–9095 (2014).
Ziolkowski, P. A. et al. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife 4, https://doi.org/10.7554/eLife.03708 (2015).
Li, X. et al. Fanconi anemia ortholog FANCM regulates meiotic crossover distribution in plants. Plant Physiol. 186, 344–360 (2021).
Blary, A. et al. FANCM Limits Meiotic Crossovers in Brassica Crops. Front. Plant Sci. 9, 368 (2018).
Mieulet, D. et al. Unleashing meiotic crossovers in crops. Nat. Plants 4, 1010–1016 (2018).
Choulet, F. et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721 (2014).
Osman, K. et al. Distal Bias of Meiotic Crossovers in Hexaploid Bread Wheat Reflects Spatio-Temporal Asymmetry of the Meiotic Program. Front. Plant Sci. 12, 631323 (2021).
Erayman, M. et al. Demarcating the gene-rich regions of the wheat genome. Nucleic Acids Res. 32, 3546–3565 (2004).
Darrier, B. et al. High-Resolution Mapping of Crossover Events in the Hexaploid Wheat Genome Suggests a Universal Recombination Mechanism. Genetics 206, 1373–1388 (2017).
Jordan, K. W. et al. The genetic architecture of genome-wide recombination rate variation in allopolyploid wheat revealed by nested association mapping. Plant J. 95, 1039–1054 (2018).
Tock, A. J. et al. Crossover-active regions of the wheat genome are distinguished by DMC1, the chromosome axis, H3K27me3, and signatures of adaptation. Genome Res. 31, 1614–1628 (2021).
Lambing, C., Franklin, F. C. & Wang, C. R. Understanding and Manipulating Meiotic Recombination in Plants. Plant Physiol. 173, 1530–1542 (2017).
Tang, Z., Yang, Z. & Fu, S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet 55, 313–318 (2014).
Chelysheva, L. et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. Plos Genet. 8, e1002799 (2012).
Morgan, C. et al. Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis. Nat. Commun. 12, 4674 (2021).
Li, W. et al. The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 138, 965–976 (2005).
Higgins, J. D. et al. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55, 28–39 (2008).
Armstrong, S. J., Caryl, A. P., Jones, G. H. & Franklin, F. C. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115, 3645–3655 (2002).
Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Jones, G. H. & Franklin, F. C. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500 (2005).
Sepsi, A., Higgins, J. D., Heslop-Harrison, J. S. & Schwarzacher, T. CENH3 morphogenesis reveals dynamic centromere associations during synaptonemal complex formation and the progression through male meiosis in hexaploid wheat. Plant J. 89, 235–249 (2016).
Appels, R. et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018).
Sandhu, R. et al. DNA Helicase Mph1(FANCM) Ensures Meiotic Recombination between Parental Chromosomes by Dissociating Precocious Displacement Loops. Dev. Cell 53, 458–472 e455 (2020).
Mancera, E., Bourgon, R., Brozzi, A., Huber, W. & Steinmetz, L. M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008).
Miao, Y. et al. Replication protein A large subunit (RPA1a) limits chiasma formation during rice meiosis. Plant Physiol. 187, 1605–1618 (2021).
Osman, K., Sanchez-Moran, E., Mann, S. C., Jones, G. H. & Franklin, F. C. Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. Embo J. 28, 394–404 (2009).
Raz, A., Dahan-Meir, T., Melamed-Bessudo, C., Leshkowitz, D. & Levy, A. A. Redistribution of Meiotic Crossovers Along Wheat Chromosomes by Virus-Induced Gene Silencing. Front Plant Sci. 11, 635139 (2020).
Arrieta, M. et al. An Induced Mutation in HvRECQL4 Increases the Overall Recombination and Restores Fertility in a Barley HvMLH3 Mutant Background. Front. Plant Sci. 12, 706560 (2021).
Maccaferri, M. et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895 (2019).
Krasileva, K. V. et al. Uncovering hidden variation in polyploid wheat. Proc. Natl Acad. Sci. USA 114, E913–E921 (2017).
Rakszegi, M. et al. Diversity of agronomic and morphological traits in a mutant population of bread wheat studied in the Healthgrain program. Euphytica 174, 409–421 (2010).
Yuan, C. et al. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6, e26468 (2011).
Desjardins, S., Kanyuka, K. & Higgins, J. D. A Cytological Analysis of Wheat Meiosis Targeted by Virus-Induced Gene Silencing (VIGS). Methods Mol. Biol. 2061, 319–330 (2020).
Peterson, R., Slovin, J. P. & Chen, C. A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 1, e13 (2010).
Higgins, J. D. Analyzing meiosis in barley. Methods Mol. Biol. 990, 135–144 (2013).
Osman, K. et al. Affinity proteomics reveals extensive phosphorylation of the Brassica chromosome axis protein ASY1 and a network of associated proteins at prophase I of meiosis. Plant J. 93, 17–33 (2018).
Mercier, R. et al. The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130, 3309–3318 (2003).
Winfield, M. O. et al. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 14, 1195–1206 (2016).
Li, Z. J. et al. The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol. 20, 139 (2019).
Guo, X. et al. De Novo Centromere Formation and Centromeric Sequence Expansion in Wheat and its Wide Hybrids. Plos Genet. 12, e1005997 (2016).
Acknowledgements
We would like to thank Neelam Dave for technical support, Helen Harper as sLola project coordinator, Claire Meade for support with crosses, and the sLola Steering Committee for helpful advice throughout the project. We thank Tobin Florio at flozbox-science for illustrating Fig. 5. This work was funded by UKRI through a BBSRC strategic Long and Large grant (sLoLa) BB/N002628/1.
Author information
Authors and Affiliations
Contributions
S.D.D., J.S., I.G., A.J.B., and K.K. performed the research; J.D.H., S.D.D., J.S., C.U., and K.J.E. designed the study; J.D.H., S.D.D., A.J.T., I.R.H., J.S., C.U., and K.J.E. analyzed the data; S.D.D., J.S., A.J.T., E.S.M., F.C.H.F., I.R.H., C.U., K.J.E., and J.D.H. wrote the paper. J.D.H. agrees to serve as the author responsible for contact and ensures communication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Marie Mirouze and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Desjardins, S.D., Simmonds, J., Guterman, I. et al. FANCM promotes class I interfering crossovers and suppresses class II non-interfering crossovers in wheat meiosis. Nat Commun 13, 3644 (2022). https://doi.org/10.1038/s41467-022-31438-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-31438-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.