Abstract
Type 3 Secretion System (T3SS) is a highly conserved virulence structure that plays an essential role in the pathogenesis of many Gram-negative pathogenic bacteria, including Pseudomonas aeruginosa. Exotoxin T (ExoT) is the only T3SS effector protein that is expressed in all T3SS-expressing P. aeruginosa strains. Here we show that T3SS recognition leads to a rapid phosphorylation cascade involving Abl / PKCδ / NLRC4, which results in NLRC4 inflammasome activation, culminating in inflammatory responses that limit P. aeruginosa infection in wounds. We further show that ExoT functions as the main anti-inflammatory agent for P. aeruginosa in that it blocks the phosphorylation cascade through Abl / PKCδ / NLRC4 by targeting CrkII, which we further demonstrate to be important for Abl transactivation and NLRC4 inflammasome activation in response to T3SS and P. aeruginosa infection.
Similar content being viewed by others
Introduction
Pseudomonas aeruginosa is one of the most virulent opportunistic bacterial pathogens, responsible for many acute and chronic-infections in wounds, the urinary tract, burns, lungs, the bloodstream, and cystic fibrosis patients, with fatality rates reaching as high as 40%1,2. P. aeruginosa derives its versatility to cause widespread infections from arsenal of cell-associated and secreted virulence factors3,4,5,6,7. Type-3 secretion-system (T3SS) virulence apparatus arguably plays the most important role in P. aeruginosa pathogenesis in human7,8. This highly conserved injectosome enables P. aeruginosa to directly translocate several effector-virulence proteins into the target-host cytoplasm where they modify host cellular processes and advance P. aeruginosa pathogenesis8,9,10,11,12,13,14. P. aeruginosa strains are generally grouped as either “cytotoxic” (expressing ExoU and ExoT), or “invasive” (expressing ExoS, ExoT, and ExoY)15,16. Without the T3SS, both cytotoxic and invasive P. aeruginosa strains become severely attenuated in their ability to cause infection8,17, highlighting the importance of T3SS and its effector proteins in P. aeruginosa pathogenesis. T3SS-expressing ExoT is the only T3SS effector maintained in all T3SS-expressing P. aeruginosa15, but what makes this virulence factor indispensable to P. aeruginosa pathogenesis remains unclear.
Prior studies have implicated both caspase-1-dependent canonical and caspase-11-dependent noncanonical inflammasomes in the recognition of and in response to P. aeruginosa infection, although caspase-11 inflammasome appears to be involved only in response to P. aeruginosa strains lacking the T3SS18,19,20,21,22. Moreover, various inflammasome subtypes (e.g., NLRP3 and/or NLRC4) have been implicated in the recognition of and in responses to T3SS and P. aeruginosa infection19,23,24,25,26,27,28. There also appears to be some contradictory reports regarding the impact of T3SS-triggered inflammatory responses on the outcome of infection, in that the same inflammasome (NLRC4) has been shown to be either crucial in combating infection and for P. aeruginosa clearance, thus benefiting the host, or paradoxically enhancing P. aeruginosa pathogenesis and facilitating bacterial colonization, thus benefiting the pathogen18,25,27,29,30,31. Further adding to the confusion in the field, ExoS and ExoU have been shown to either dampen pro-inflammatory responses or trigger pro-inflammatory responses25,32,33,34,35,36,37,38. These reports suggest that specific organs/sites within a host may have evolved distinct mechanism(s) to detect and respond to T3SS and its effectors during P. aeruginosa infection. Of note, the impact of ExoT on inflammatory responses within the host has not been directly evaluated.
Wound is a favorite niche for P. aeruginosa infections39,40,41,42. P. aeruginosa is the number-one Gram-negative pathogen detected in both chronic and acute wounds, and its presence in wounds correlates with a poor prognosis for healing in human43,44,45,46. In line with these reports, P. aeruginosa has been shown to employ various virulence strategies to alter cellular functions and to inhibit wound healing as a way to expand its favorite niche47,48,49,50,51,52. Of note, T3SS plays a pivotal role in inhibiting tissue repair and exacerbating wound damage47,48, highlighting the importance of the T3SS function to P. aeruginosa pathogenesis in wounds.
In this report, we examined the impact of P. aeruginosa on host immune responses within the wound environment, with the focus on the T3SS and ExoT, and determined which interactions may benefit the host and which may benefit P. aeruginosa in this evolutionary tug of war. Our data demonstrate that recognition of P. aeruginosa T3SS triggers a phosphorylation cascade through the Abl/PKCδ/NLRC4, culminating in NLRC4 inflammasome activation and inflammatory responses that limit P. aeruginosa infection in wounds. We further show that P. aeruginosa ExoT dampens the phosphorylation cascade through Abl/PKCδ/NLRC4 and inhibits NLRC4 inflammasome activation by targeting CrkII, which we further demonstrate to be important for Abl transactivation and NLRC4 inflammasome activation in response to T3SS and P. aeruginosa infection.
Results
ExoT dampens pro-inflammatory cytokine production in wound
We used PA103—(a representative cytotoxic strain7,8,53), and PAK—(a representative invasive strain7,8), and their isogenic ExoT, ExoS, and ExoU T3SS mutant strains in a full-thickness excisional wound in C57BL/6 mice, as described previously47,54. Wounds were infected with 103 wild-type or T3SS mutant strains and the impact of infection with wild-type and T3SS mutants on the production of IL-1β and IL-18 pro-inflammatory chemokines (products of inflammasomes and important pro-inflammatory cytokines against P. aeruginosa infection55,56) was assessed by enzyme-linked immunosorbent assay (ELISA) 24 h after infection.
Infection with PA103 or PAK wild-type strains resulted in significantly more IL-1β in wounds as compared with PBS-treated uninfected wounds (Fig. 1a, Supplementary Fig. 1a, c, & e).
a–g C57BL/6 wound tissues were harvested 24 h after treatment with PBS or infection with 103 wildtype P. aeruginosa PA103 and the indicated T3SS isogenic mutants. a, b About 24 h after infection, tissue homogenates were assessed for the pro-inflammatory cytokines IL-1β (a) or IL-18 (b) by ELISA, and the tabulated data are shown as the mean ± SEM. (N = 8 mice/group; 2 wounds/mouse). c, d Wounds were fixed and assessed for leukocyte migration by H&E staining. Representative images in the dermal regions from underneath the wounds are shown in (c) and the corresponding tabulated number of leukocytes is shown as the mean ± SEM in (d) (N = 5 mice/group; ~7 random fields/wound/mouse). e, f Neutrophil activation was assessed by determining MPO positive cells by immunohistochemistry assay (IHC). Representative images in the dermal regions from underneath the wounds are shown in (e) and the corresponding tabulated number of MPO-positive neutrophils is shown as the mean ± SEM in (f) (N = 5 mice/group, ~7 random fields/wound/mouse). g Tissue homogenates from day 1 wounds were assessed for MPO levels by ELISA (N = 5 mice/group). Statistical analyses were determined by one-way ANOVA with post hoc testing. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Exact P-values are presented in Supplementary Data 1. Source data are provided as a Source Data file.
IL-18 was also increased in PAK-infected wounds, but not significantly increased in PA103-infected wounds (Fig. 1b & Supplementary Fig. 1b & d). Intriguingly, regardless of the strain genetic background, wounds infected with ExoT-deleted (∆T) strains, contained substantially more IL-1β and IL-18 as compared with their isogenic wildtype strains, and complementing these ∆T strains with a plasmid expressing ExoT (pExoT) reduced IL-1β and IL-18 levels significantly, suggesting that ExoT dampens inflammatory responses in wound, at least with respect to IL-1β and IL-18 inflammatory cytokines (Fig. 1a–b and Supplementary Fig. 1a–e). In contrast, wounds infected with ExoS-deleted (∆S) or ExoU-deleted (∆U) strains contained similar levels of IL-1β and IL-18 as compared with their wild-type counterparts, indicating that in wound, ExoS and ExoU do not possess anti- or pro-inflammatory functions.
ExoT dampens leukocyte migration and activation in wound
We next assessed leukocyte infiltration in wounds treated with PBS or infected with the aforementioned P. aeruginosa strains, by hematoxylin and eosin (H&E) histological analysis, which is an established method for this purpose47,54,57,58,59,60. Consistent with the IL-1β and IL-18 chemokine levels (Fig. 1a–b and Supplementary Fig. 1a–e), wounds infected with ExoT-deleted mutant strains also contained significantly more leukocytes as compared with wild-type strains or ExoT-complemented strains (Fig. 1c–d, and Supplementary Fig. 2a–b). Corroborating these data, assessment of activated neutrophil contents in the aforementioned wounds by histological analysis of myeloperoxidase (MPO)-positive cells or by MPO measurements by ELISA also indicated significantly higher levels of activated neutrophils in wounds that were infected with ExoT-deleted mutant strains as compared with wild-type or ExoT-complemented strains (Fig. 1e–g and Supplementary Fig. 2c–d). (MPO is primarily a marker for activated neutrophils54,58,61).
NLRC4 is the primary inflammasome responsible for P. aeruginosa recognition and it is inhibited by ExoT
ExoS and ExoU have been previously reported to dampen IL-1β production25,32, but the impact of ExoT on inflammasome activity in bone marrow-derived macrophages (BMDMs) has not been directly examined. Since our in vivo data clearly demonstrated that ExoT (not ExoS or ExoU) is the T3SS effector-virulence factor that dampens the production of IL-1β and IL-18 in wound (Fig. 1, Supplementary Fig. 1a–e, & Supplementary Fig. 2), we wished to assess the impact of ExoT on inflammasome activity in BMDMs. Toward this goal, we extracted BMDMs from C57BL/6 normal mice and infected them with wild-type or ExoT-deleted P. aeruginosa strains for 2 h. In line with our in vivo results and regardless of strain genetic background, infection with ∆T mutant strains resulted in significantly higher levels of IL-1β and IL-18 secreted by BMDMs, and complementing ∆T strains with pExoT reduced the production of these pro-inflammatory cytokines (Fig. 2a–b and Supplementary Fig. 3a–b).
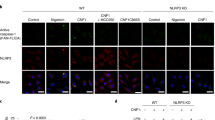
a–j BMDMs (from C57BL/6) were infected with wild-type P. aeruginosa PA103 and the indicated T3SS isogenic mutants for 1-2 h (1 h for IF microscopy and 2 h for ELISA and Western blotting). a, b Culture supernatants were assessed for IL-1β or IL-18 by ELISA and the tabulated data are shown as the mean ± SEM. (N = 3; ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). c, d BMDMs of caspase-1 or Caspase-11-knockout mice (Cas-1−/− or Cas-11−/−) were infected with PA103. Culture supernatants were assessed for IL-1β or IL-18 by ELISA and the tabulated data are shown as the mean ± SEM. (N = 4; ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). e, f Cell lysates and supernatants were assessed for caspase-1 activation by Western blotting (e) and the corresponding densitometer data are shown as the mean ± SEM in (f) (N = 3, each experiment in e & f was repeated independently 2 times. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (g, h) BMDMs of indicated inflammasome knockout mice were infected with PA103. Culture supernatants were assessed for IL-1β or IL-18 by ELISA and the tabulated data are shown as the mean ± SEM (N = 4; ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (i–j) BMDMs (from C57BL/6) were infected with PA103 and the indicated T3SS isogenic mutants. They were then fixed and stained for p-NLRC4 (green), ASC (red), and nucleus/DAPI (blue). Colocalized p-NLRC4/ASC foci were assessed by IF microscopy. Representative images are shown in (i), and the tabulated data are shown as the mean ± SEM in (j) (N = 3 replicates, ≥7 random fields per replicate. ns, not significant; p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Arrows point to representative p-NLRC4/ASC foci within BMDMs. k–n Wounds of C57BL/6, ASC, Caspase-1 or NLRC4 knockout mice were infected with 103 of PA103 and the indicated T3SS isogenic mutants. Bacterial burden in wounds were determined by serial dilution and plating, 24 h after infection and the tabulated data are shown as the mean ± SEM. (N = 5 mice/group for C57BL/6, N = 4 mice/group for ASC and NLRC4 knockout mice and N = 8 mice/group for Caspase-1 knockout mice. Statistical analyses were determined by one-way ANOVA with post hoc testing; ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Exact P values are presented in Supplementary Data 1. Source data are provided as a Source Data file.
Although, most studies point to the caspase-1-dependent canonical inflammasomes in mounting inflammatory responses to P. aeruginosa infection18,19, caspase-11 dependent noncanonical inflammasome has also been implicated in inflammatory responses to P. aeruginosa20,21,22,62,63,64, prompting us to reassess the role of canonical and noncanonical inflammasomes in the recognition of and mounting inflammatory responses to P. aeruginosa infection. Toward this goal, we challenged BMDMs extracted from C57BL/6 (WT), or caspase-1 or caspase-11 knockout mice (Cas-1–/– and Cas-11–/–, respectively), with the P. aeruginosa. Culture supernatants were then examined for the production of IL-1β and IL-18 by ELISA. Data indicated that while BMDMs from WT or Cas-11–/– mice recognized and responded to P. aeruginosa infection, Cas-1–/– BMDMs were impaired in responding to P. aeruginosa infection, as manifested by significant reductions in IL-1β and IL-18 production (Fig. 2c–d), suggesting that P. aeruginosa recognition in BMDMs is primarily mediated by caspase-1 dependent canonical inflammasome. In line with ExoT’s anti-inflammatory function in wound, infection with ExoT-deleted P. aeruginosa strains also resulted in substantial increases in caspase-1 activation as assessed by Western immunoblotting probing for the processed active caspase-1 fragment (p20) in BMDMs, and caspase-1 inflammasome assembly as determined by caspase-1/ASC foci number determination by immunofluorescent (IF) microscopy65 (Fig. 2e–f and Supplementary Fig. 3c–d).
NLRP1b, NLRP3, NLRC4, AIM2, and Pyrin are the five well- characterized subtypes of caspase-1-dependent canonical inflammasomes66,67. Depending on the host and/or cell line, P. aeruginosa has been shown to activate NLRC4 or NLRP3 inflammasomes25,26,27,28,68, suggesting that eukaryotic hosts may utilize different caspase-1 inflammasome subtypes to mount inflammatory responses to P. aeruginosa infection. To determine which caspase-1-dependent canonical inflammasome subtype(s) contributed to P. aeruginosa recognition and response in BMDMs, we extracted BMDMs from knockout mice defective in these inflammasomes (Nlrp1–/–, Nlrp3–/–, Nlrc4–/–, Aim2–/–, and Pyrin–/–) and infected them with P. aeruginosa and assessed their supernatants for the production of IL-1β and IL-18 by ELISA. Data indicated that NLRC4 was the response in BMDMs, as defect in this inflammasome reduced IL-1β and IL-18 production to levels comparable to Cas-1–/– BMDMs (Fig. 2g–h and Supplementary Fig. 3e). Moreover, infection with ExoT-deleted mutant strains resulted in nearly 3-fold increase in the number of NLRC4 inflammasome foci and complementing this strain with pExoT, but not empty vector, significantly reduced NLRC4 inflammasome foci numbers in BMDMs (Fig. 2i–j), indicating that ExoT interferes with NLRC4 inflammasome assembly.
ExoT enhances bacterial fitness in the wounds by dampening the NLRC4 inflammasome
We next determined bacteria (infection) burden in the wounds infected with the wildtype or T3SS mutant strains by colony-forming unit (CFU) determination, as described47,54,69,70. Data indicated that regardless of genetic background, ExoT-deleted mutant strains were significantly impaired in their ability to colonize wound, as manifested by approximately 1–2 log-order reductions in bacterial counts in wound as compared with the wild-type strains (Fig. 2k and Supplementary Fig. 3f). Complementing ∆T mutant strains with pExoT, (but not with the empty vector pUCP20), restored their ability to colonize the wound, suggesting that heightened inflammatory responses in wounds are responsible for the reduction in the fitness of ExoT-deleted mutant strains in this environment. Of note, ExoS-deleted mutants also exhibited reduced colonization in wounds, but ExoU-deleted strains did not, indicating that ExoS (but not ExoU), imparts additional fitness benefit to P. aeruginosa in wound. In line with this notion, PAK strain consistently colonized wounds ~1 log order more than PA103 strain (Supplementary Fig. 3h), suggesting that PAK’s enhanced colonization in wound may be due to its possession of ExoS virulence factor.
To assess the role of caspase-1-dependent NLRC4 inflammasome function in controlling P. aeruginosa infection in wound, we challenged wounds in mice deficient in caspase-1 (Cas-1–/–), or the adaptor protein ASC (ASC–/–), which is required for all caspase-1 dependent canonical inflammasome subtypes71, or NLRC4 (Nlrc4–/–), with the aforementioned P. aeruginosa strains. Regardless of strain genetic background, wounds in Cas-1–/–, ASC–/–, and Nlrc4–/– mice were significantly more vulnerable to infection with P. aeruginosa as they contained >1 log-order more bacteria than wounds in C57BL/6 normal mice (Fig. 2k–n, and Supplementary Fig. 3f–g). Moreover, the presence or absence of T3SS or ExoT did not impact the ability of P. aeruginosa to colonize wounds in caspase-1- or NLRC4 inflammasome defective mice, indicating that the primary virulence function of ExoT in wound is to dampen inflammatory responses through NLRC4 inflammasome (Fig. 2k–n, and Supplementary Fig. 3f–g).
P. aeruginosa infection in C57BL/6 wounds peaks on day 3, but begins to resolve by day 647. We conducted a similar time-course infection study in normal and NLRC4−/− mice to assess the role of NLRC4 inflammasome in combating P. aeruginosa infection. Data indicated that NLRC4−/− wounds contained significantly more bacteria than normal wounds on days 1, 3, and 6, highlighting the importance of NLRC4 in combating P. aeruginosa infection in wound (Supplementary Fig. 3i). Collectively, these data indicated that T3SS-expressing P. aeruginosa recognition in wound by NLRC4 canonical inflammasome results in pro-inflammatory responses that limit P. aeruginosa infection. The data further indicated that ExoT functions as the main anti-inflammatory agent for P. aeruginosa, providing a necessary stealth mechanism against the host innate immune responses for this pathogen in wound environment.
NLRC4 phosphorylation by PKCδ plays a critical role for NLRC4 inflammasome activation in response to P. aeruginosa infection
There appears to be some controversy regarding the role of NLRC4 phosphorylation at serine residue 533 (Ser 533) in NLRC4 inflammasome activity, with some groups reporting it to be essential for NLRC4 inflammasome activation72,73,74, while others questioning the dependence of NLRC4 inflammasome activity on the NLRC4 phosphorylation75,76. To evaluate the dependence of NLRC4 inflammasome activation on NLRC4 phosphorylation in response to P. aeruginosa infection, we reconstituted NLRC4−/− BMDMs with either wild-type NLRC4 or nonphosphorylatable NLRC4 mutant (NLRC4/S533A)72 (Methods), and assessed their responses to P. aeruginosa infection. Data indicated that NLRC4/S533A-expressing BMDMs were significantly impaired in responding to P. aeruginosa infection as assessed by Western blotting, probing for activated caspase-1 (p20), and IL-1β and IL-18 production (Fig. 3a–c, and Supplementary Fig. 4a). These data indicated that NLRC4 phosphorylation is required for NLRC4 inflammasome activation in response to P. aeruginosa infection.
a–c Nlrc4−/− BMDMs were reconstituted with wildtype NLRC4 or nonphosphorylatable NLRC4 mutant (NLRC4/S533A). They were then infected with P. aeruginosa ΔUΔT (T3SS+) strain for 2 h. Cell lysates and supernatants were assessed for caspase-1 activation by Western blotting (a) and culture supernatants were assessed for IL-1β or IL-18 by ELISA and the tabulated data are shown as the Mean ± SEM in (b, c) respectively. d–f BMDMs from C57BL/6 were transfected for PKCδ siRNA for 48 h. After transfection, cells were infected with P. aeruginosa ΔUΔT for 2 h. Cell lysates were assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting (d) and culture supernatants were assessed for IL-1β or IL-18 cytokines by ELISA, and the tabulated data are shown as the mean ± SEM in (e, f) respectively. g–i BMDMs from C57BL/6 were pretreated with PKCδ inhibitor Rottlerin (5 µM) for 90 min and then cells were infected with P. aeruginosa ΔUΔT for 2 h. Whole-cell lysate was assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting (g) and culture supernatants were assessed for IL-1β and IL-18 cytokines by ELISA and the tabulated data are shown as the mean ± SEM (h, i), respectively. j BMDMs (from C57BL/6 and Nlrc4−/−) were infected with ΔUΔT for 2 h. Cell lysates were assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting. (Statistical analyses were determined by one-way ANOVA with post hoc testing. Each experiment was repeated independently three times, except (a), which was repeated twice (N = 3 each time). ns, not significant; * p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Exact P-values are presented in Supplementary Data 1. Source data are provided as a Source Data file.
NLRC4 phosphorylation at Ser 533 is mediated by PKCδ kinase and has been shown to be essential for NLRC4 inflammasome activity in BMDMs infected with Salmonella enterica serovar Typhimurium (a.k.a., Salmonella Typhimurium)72. However, the role of PKCδ in NLRC4 inflammasome activation has also been questioned75,76, prompting us to assess the dependence of NLRC4 inflammasome activity on PKCδ kinase activity in response to P. aeruginosa. PKCδ protein depletion by treatment with PKCδ-specific siRNA77, or pretreatment of BMDMs with PKCδ kinase inhibitor Rottlerin72, significantly reduced NLRC4 phosphorylation, caspase-1 activation, and IL-1β and IL-18 production, in BMDMs infected with P. aeruginosa (Fig. 3d–i and Supplementary Fig. 4b–c). Of note, while caspase-1 processing/activation was substantially reduced in NLRC4−/− BMDMs infected with P. aeruginosa, PKCδ phosphorylation was not affected, indicating that PKCδ is upstream of NLRC4 (Fig. 3j and Supplementary Fig. 4d). These results indicated that PKCδ kinase activity is essential for NLRC4 phosphorylation and NLRC4 inflammasome activation in response to P. aeruginosa.
NLRC4 inflammasome inhibition by ExoT is primarily dependent on its ADPRT-domain activity
ExoT is a bifunctional virulence factor that contains a GTPase-activating protein (GAP) domain at its N-terminus and an ADP-ribosyltransferase (ADPRT) domain at its C-terminus78,79. To gain insight into the mechanism by which ExoT interferes with NLRC4 inflammasome activity, we assessed the contributions of the GAP and the ADPRT domains in mediating ExoT’s anti-inflammasome function in BMDMs. Data indicated that ExoT—in a manner that was primarily dependent on its ADPRT-domain activity—dampened PKCδ and NLRC4 phosphorylation, inhibited caspase-1 processing, interfered with IL-1β and IL-18 production, and interfered with caspase-1 inflammasome assembly (Fig. 4a–f). In line with these results, ExoT’s anti-inflammatory function in wound was also mediated by its ADPRT-domain activity, as mutation in the ADPRT domain activity abrogated ExoT’s ability to dampen IL-1β production or enhance P. aeruginosa colonization in wound (Supplementary Fig. 5a–b). However, the underlying mechanism responsible for inhibition of NLRC4 inflammasome by ExoT/ADPRT domain remained unclear.
a, b BMDMs from C57BL/6 were infected with indicated P. aeruginosa strains for 2 h. Supernatants and cell lysates were then assessed for indicated phosphorylated and unphosphorylated proteins by Western blotting (a), and the corresponding densitometer data are shown in (b) as the mean ± SD. (Each experiment in a–b was repeated independently at least two times) (N = 3, ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). c, d IL-1β and Il-18 cytokines were assessed by ELISA and the tabulated data are shown as the mean ± SD. (Each experiment was done twice independently) (N = 3 replicates each time). ns, not significant; p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. e, f Colocalized caspase-1/ASC foci were assessed by IF microscopy. Representative images are shown in (e) and the tabulated data are shown as the mean ± SD in (f). Arrows point to some caspase-1/ASC foci within BMDMs. Insets are the magnified images of representative caspase-1/ASC foci. The scale bars are 5 µm. (N = 3 replicates; ≥7 random fields per replicate. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical analyses were determined by one-way ANOVA with post hoc testing). Exact P-values are presented in Supplementary Data 1. Source data are provided as a Source Data file.
ExoT/ADPRT blocks NLRC4 inflammasome by dampening the phosphorylation cascade through NLRC4 inflammasome by targeting Crk adaptor protein
PKCδ itself becomes phosphorylated at tyrosine residue 311 (Tyr 311) and PKCδ phosphorylation has been shown to be required for NLRC4 phosphorylation and NLRC4 inflammasome activation in BMDMs in response to infection with Salmonella Typhimurium72, but the identity of the tyrosine kinase responsible for PKCδ phosphorylation has remained unknown for nearly a decade. ExoT/ADPRT domain has been demonstrated to ADP-ribosylate and inactivate Crk adaptor protein9,10,52,79. Interestingly, CrkI and CrkII isoforms of Crk adaptor protein were originally identified by their homology to the oncogene product of avian sarcoma virus CT10 (v-Crk)—which despite the absence of an apparent catalytic domain in the oncogene—induces massive increases in tyrosine-phosphorylated proteins in chicken-embryo fibroblasts transformed by this virus80,81. We posited that ExoT may inhibit P. aeruginosa-triggered NLRC4 activation by targeting Crk adaptor protein that we further postulated to be essential (either directly or indirectly) for PKCδ phosphorylation and for NLRC4 inflammasome activation.
To test our hypothesis, we first evaluated the dependence of PKCδ and NLRC4 phosphorylation and NLRC4 inflammasome activation on Crk in response to P. aeruginosa, by depleting BMDMs of Crk adaptor protein by Crk-specific siRNA9,82 prior to infection with P. aeruginosa. Treatment with Crk siRNA reduced the levels of CrkI and CrkII isoforms by >90%, and significantly reduced PKCδ and NLRC4 phosphorylation, and caspase-1 processing as assessed by Western blotting (Fig. 5a–b), and significantly dampened NLRC4 inflammasome activity as assessed by IL-1β and IL-18 analysis by ELISA (Fig. 5c–d). These data indicated that Crk adaptor protein plays a significant role in PKCδ and NLRC4 phosphorylation and for NLRC4 inflammasome activation in response to P. aeruginosa infection, but how Crk mediated NLRC4 inflammasome activation in response to P. aeruginosa infection remained unclear.
a–d BMDMs from C57BL/6 were transfected for Crk siRNA for 48 h. After transfection, cells were infected with P. aeruginosa ΔUΔT strain for 2 h. Whole-cell lysate was assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting (a), and the corresponding densitometer data are shown as the mean ± SEM in (b). (Each experiment in a, b was repeated at least three times independently). c, d Culture supernatants were assessed for IL-1β and IL-18 cytokines by ELISA and the tabulated data are shown as the Mean ± SEM. (Each experiment was repeated at least 3 times independently with N = 3 replicates each time; ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Statistical analyses were determined by one-way ANOVA with post hoc testing. Exact P-values are presented in Supplementary Data 1. Source data are provided as a Source Data file.
Crk-dependent Abl tyrosine-kinase activity is required for PKCδ phosphorylation and NLRC4 inflammasome activation in response to P. aeruginosa infection
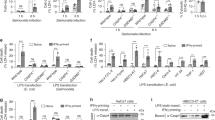
To hone in on the role of Crk in NLRC4 inflammasome activity, we next evaluated the interaction between Crk and PKCδ and NLRC4 by co-immunoprecipitation (Co-IP)-pulldown assay, using PKCδ or NLRC4 as baits. While PKCδ and NLRC4 co-immunoprecipitated with each other in response to infection as expected, neither protein co-immunoprecipitated with CrkI or CrkII isoforms of Crk (Fig. 6a and Supplementary Fig. 6a), suggesting that Crk functions upstream of PKCδ phosphorylation. FAK, Src, Lck, Fyn, Lyn, and Abl are all tyrosine kinases, which have been shown to either directly or indirectly interact with Crk under various conditions83,84,85,86,87,88. We wondered whether one of these tyrosine kinases may be responsible for PKCδ phosphorylation and NLRC4 activation in response to P. aeruginosa infection. To test our hypothesis, we infected BMDMs with P. aeruginosa after pretreating them with specific inhibitors of the aforementioned tyrosine kinases89,90,91,92,93,94,95. Data indicated that pretreatment with Abl inhibitor (Imatinib) significantly reduced IL-1β production in BMDMs infected with ExoT-deficient P. aeruginosa (Fig. 6b). Corroborating these data and ruling out off-target effects of Imatinib, Abl-depleted BMDMs, after treatment with Abl-specific siRNA (96 and Methods), and Abl-knockout BMDMs, extracted from Ablflox/flox LysM Cre + mice (97 and Methods), also displayed significant reductions in PKCδ and NLRC4 phosphorylation, caspase-1 activation, and IL-1β and IL-18 production in BMDMs infected with P. aeruginosa (Fig. 6c–h, and Supplementary Fig. 6b–c).
a BMDMs from C57BL/6 were infected with the indicated P. aeruginosa strain for 2 h. Whole-cell lysates were immunoprecipitated with antibodies against NLRC4, PKCδ or Abl and immunoblotted for indicated proteins. b BMDMs from C57BL/6 were pretreated with inhibitors of indicated Crk-interacting tyrosine kinases (Methods) for 90 min and then infected with ΔUΔT for 2 h, and the supernatants were assessed for IL-1β and IL-18 by ELISA and the tabulated data are shown as the mean ± SEM. c–e BMDMs from C57BL/6 were transfected with Abl siRNA for 24 h. After transfection, cells were either left uninfected (media) or infected with the indicated P. aeruginosa strain for 2 h. Whole-cell lysates were assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting (c), and culture supernatants were assessed for IL-1β and IL-18 by ELISA, and the tabulated data are shown as the mean ± SEM in (d, e), respectively. f–h BMDMs from Ablflox/flox LysM Cre- and Ablflox/flox LysM Cre + mice were infected with the indicated P. aeruginosa strain for 2 h. Whole-cell lysates were assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting (f), and culture supernatants were assessed for IL-1β and IL-18 cytokines by ELISA and the tabulated data are shown as the mean ± SEM in (g, h), respectively. i BMDMs from C57BL/6 were infected with indicated P. aeruginosa strains for 2 h. Whole cell lysates were assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting. j, k BMDMs from C57BL/6 were transfected with Crk siRNA for 48 h. After transfection, cells were infected with indicated P. aeruginosa strain for 2 h. Whole cell lysate was assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting (j) and the corresponding densitometer data are shown as the mean ± SEM in (k). l BMDMs from C57BL/6 were infected with indicated P. aeruginosa strains for 2 h. Whole-cell lysates and supernatants were immunoprecipitated with antibodies against NLRC4, PKCδ, Crk or Abl and immunoblotted for indicated proteins. (Experiments were repeated twice independently, except (c–e), which were repeated at least 3 times independently with N = 3 replicates each time. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical analyses were determined by one-way ANOVA with post hoc testing for all except (g, h), which were done by two-sided unpaired Student’s t-test). Exact P-values are presented in Supplementary Data 1. Source data are provided as a Source Data file.
Intriguingly, Abl co-immunoprecipitated with PKCδ but not with NLRC4 in response to infection with P. aeruginosa (Fig. 6a and Supplementary Fig. 6a). Moreover, Abl phosphorylation in response to P. aeruginosa infection was not affected in BMDMs pretreated with PKCδ inhibitor Rottlerin72 or Nlrc4–/– or in Cas-1–/– BMDMs, while PKCδ inhibition abrogated NLRC4 phosphorylation and caspase-1 activation in response to P. aeruginosa infection (Supplementary Fig. 7a–d, Fig. 3d–j, and Supplementary Fig. 4b–c). These data further support the notion of Abl functioning upstream of PKCδ, mediating its phosphorylation, and PKCδ functioning upstream of NLRC4, mediating its phosphorylation in response to infection with P. aeruginosa.
Abl-kinase interaction with Crk adaptor protein has been shown to result in Abl activation by autophosphorylation (transactivation) at tyrosine 245 (Tyr 245)98,99. Intriguingly, Abl also co-immunoprecipitated with CrkII isoform of Crk in response to P. aeruginosa infection (Fig. 6a and Supplementary Fig. 6), suggesting that Abl interaction with CrkII may be important for Abl transactivation in BMDMs in response to P. aeruginosa infection. To address this possibility, we first assessed whether Abl becomes phosphorylated at Tyr 245 in BMDMs in response to P. aeruginosa. Infection with P. aeruginosa strain resulted in substantial increases in Abl phosphorylation at Tyr 245, which was significantly inhibited by ExoT/ADPRT (Fig. 6i and Supplementary Fig. 6d). Consistent with Crk’s role in Abl transactivation98,99, Crk-protein depletion by siRNA led to ~90% reduction in Abl phosphorylation in BMDMs infected with P. aeruginosa (Fig. 6j–k). These data indicated that Abl transactivation in response to P. aeruginosa infection is dependent on its interaction with Crk adaptor protein, and suggested that Crk/Abl interaction is disrupted by ExoT/ADPRT.
To assess the interactions between ExoT with Crk and/or Abl, we transfected BMDMs with a mammalian expression vector, harboring ExoT or inactive mutant ExoT (ExoT G−A−), fused to GFP at their C-termini, in the presence of pancaspase inhibitor Z-VAD to block ExoT/GAP and ExoT/ADPRT-induced apoptosis10,100,101. (These vectors have been described previously and GFP fusion does not alter ExoT activities9,10,52,100,102). Data demonstrated that ExoT-GFP and ExoT(G−A−)–GFP both interacted with CrkI and CrkII isoforms of Crk but not with Abl, as assessed by Co-IP, using GFP as the bait (Supplementary Fig. 8a–b), indicating that ExoT does not directly interact with Abl. Given that CrkII only interacted with Abl in response to infection (Fig. 6a), we next infected the aforementioned transfected BMDMs with ∆U∆T P. aeruginosa strain and assessed the interaction between ExoT–GFP and ExoT(G−A−)–GFP with Crk and Abl in the aforementioned transfected BMDMs under infection. ExoT or ExoT(G−A−) co-immunoprecipitated with both CrkI and CrkII in response to infection (Supplementary Fig. 8c–d). In contrast, Abl co-immunoprecipitated with ExoT(G−A−) but significantly less with ExoT, suggesting that Abl interaction with ExoT (G−A−) is likely indirect through CrkII and ExoT interferes with CrkII/Abl interaction (Supplementary Fig. 8c–d).
To further explore this possibility, we next assessed the impact of ExoT on CrkII/Abl interaction by Co-IP pull-down assay in BMDMs infected with ExoT-deleted or ExoT-expressing strains. Data indicated that ExoT disrupted CrkII/Abl interaction in a manner that was primarily dependent on its ADPRT-domain activity (Fig. 6l and Supplementary Fig. 9). These data are consistent with ExoT’s inhibition of Abl transactivation that is dependent on its interaction with CrkII (Fig. 6i–k). Of note, interactions between Abl/PKCδ and PKCδ/NLRC4 were also significantly disrupted by ExoT in a manner that was dependent on its ADPRT activity, indicating that these interactions are dependent on CrkII/Abl interaction and Abl transactivation (Fig. 6l and Supplementary Fig. 9).
We next assessed the involvement of Abl and PKCδ in caspase-1 inflammasome activity toward WI-14 and WI-16 P. aeruginosa wound clinical isolates, which are respectively similar to PA103 and PAK strains in regard to their T3SS effectors15. Infection with these clinical strains also led to caspase-1 activation in BMDMs and significant increases in IL-1β and IL-18 production (Supplementary Fig. 10a–d). Moreover, Abl and PKCδ protein depletion by their specific siRNAs also significantly dampened IL-1β and IL-18 production in BMDMs infected with these clinical isolates (Supplementary Fig. 10e–g), further highlighting the importance of Abl and PKCδ in inflammatory responses to T3SS-expressing P. aeruginosa.
Abl kinase is required for NLRC4 inflammasome activation in response to P. aeruginosa infection in wound
To assess the importance of Abl kinase function in NLRC4 inflammasome activation in response to P. aeruginosa infection in wound, we first assessed the phosphorylation status of Abl, PKCδ, and NLRC4 in wound infected with ExoT-deleted or ExoT-expressing P. aeruginosa strains. Data demonstrated that wounds infected with wild-type P. aeruginosa contained significantly higher levels of phosphorylated Abl, PKCδ, and NLRC4, and activated caspase-1 (p20) as compared with uninfected wounds (Fig. 7a–b), indicating that NLRC4 inflammasome becomes activated in wound in response to P. aeruginosa infection. Consistent with the in vitro data in BMDMs, wounds infected with ExoT-deleted mutants contained significantly higher levels of phosphorylated Abl, PKCδ, and NLRC4, and activated caspase-1 (p20), as compared with wounds infected with wild-type P. aeruginosa strain or ExoT-deleted strain complemented with ExoT (Fig. 7a–b), indicating that ExoT dampens NLRC4 inflammasome activation in wound.
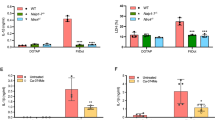
a, b C57BL/6 wound tissues were harvested 24 h after treatment with PBS or infection with 103 WT PA103, and ExoT-deleted (∆T) isogenic strains with or without ExoT complementation. Tissue homogenates were assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting and the corresponding densitometer data are shown as the mean ± SEM (N = 3). Statistical analyses were determined by one-way ANOVA with post hoc testing, ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. c–i C57Bl/6 mice received Imatinib (100 mg/kg) or PBS (mock) by i.p injection (Materials). Wounds were infected with 103 of P. aeruginosa ∆U∆T strain. After 24 h of infection, tissue homogenates were assessed for indicated proteins by Western blotting (c) and for the pro-inflammatory cytokines by ELISA, and the tabulated data are shown as the mean ± SEM (d, e) (N = 8). (f, g) Wounds were fixed and assessed for leukocyte migration by H&E staining. Representative images in the dermal regions from underneath the wounds are shown in (f) and the corresponding tabulated number of leukocytes is shown as the mean ± SEM in (g) (N = 5 mice/group, ~7 random fields/wound/mouse). h Tissue homogenates from day-1 wounds were assessed for MPO levels by ELISA. i Bacterial burden was determined by serial dilution and plating after 24 h of infection and the tabulated data are shown as the mean ± SEM (N = 8; ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical analyses were determined by two-sided unpaired Student t-test). Exact P-values are presented in Supplementary Data 1. Source data are provided as a Source Data file.
Abl deletion causes lethality at embryonic day 8–9 because of Abl’s crucial roles in vascular function and in development103,104. To assess the role of Abl kinase in NLRC4 inflammasome activity in response to P. aeruginosa infection in wound, we treated C57BL/6 mice with Abl inhibitor Imatinib or PBS by intraperitoneal injection, prior to wounding and infection with P. aeruginosa (Methods). We next assessed the wounds for inflammatory responses and infection burden 24 h after infection. As expected, wounds in Imatinib-treated mice contained significantly lower levels of phosphorylated Abl (Fig. 7c and Supplementary Fig. 11). Consistent with the role of Abl kinase in PKCδ phosphorylation and NLRC4 inflammasome activation in response to P. aeruginosa (Fig. 6b–h and Supplementary Fig. 6a–c), Imatinib-treated wounds also contained significantly lower levels of phosphorylated PKCδ and NLRC4, and activated caspase-1 (p20) as assessed by Western blotting (Fig. 7c and Supplementary Fig. 11); reduced IL-1β and IL-18, as assessed by ELISA (Fig. 7d–e); reduced leukocyte content as assessed by H&E staining (Fig. 7f–g); and reduced activated neutrophils as assessed by MPO analysis by ELISA (Fig. 7h). Importantly, Imatinib-treated mice contained nearly 2 log-order more bacteria in their wounds, as compared with mock-treated mice (Fig. 7i). Collectively, these data indicated that Abl function is essential for NLRC4 inflammasome activation and in combating P. aeruginosa infection in wound.
Discussion
In this report, we assessed the impact of P. aeruginosa infection on inflammatory responses within the wound environment. Our data show that recognition of T3SS-expressing P. aeruginosa strains by the caspase-1 dependent canonical inflammasome leads to pro-inflammatory responses, manifested by increases in caspase-1-activation, IL-1β, and IL-18 cytokine production, and leukocyte migration to the site of infection. Our data further indicate that NLRC4 canonical inflammasome subtype is primarily responsible for the recognition of and response to P. aeruginosa infection in BMDMs and in wound. Interestingly, NLRC4 inflammasome has been shown to either protect the host against P. aeruginosa infection or paradoxically facilitate and enhance P. aeruginosa colonization and pathogenesis18,31. Our data indicate that in wound, NLRC4 inflammasome activity provides crucial protection against P. aeruginosa infection, as wounds in mice defective in the components of NLRC4 inflammasome are highly vulnerable to P. aeruginosa infection.
There appears to be some controversy regarding the role of PKCδ kinase and NLRC4 phosphorylation in NLRC4 inflammasome activity, with some groups reporting them to be essential72,73,74, while others disputing their role in NLRC4 inflammasome activity75,76. These seeming differences may reflect the different microorganisms used in these studies (e.g., Salmonella, Shigella, and Legionella); the different experimental procedures used for analyses (e.g., BMDM infection with bacteria, vs. delivering bacterial products, such as flagellin by FlaTox, vs. in vitro reconstitution assays); and the complexity of inflammasome responses emerging when multiple factors (e.g., T3SS, T4SS, or flagellin) are analyzed independently or in combination, as have been the case in these studies. Our data clearly demonstrate that NLRC4 phosphorylation by PKCδ is essential for NLRC4 activation and pro-inflammatory responses to P. aeruginosa infection. We show that NLRC4 is phosphorylated in response to P. aeruginosa, both in vitro and in vivo. We further show that BMDMs expressing nonphosphorylatable NLRC4 are impaired in their ability to respond to P. aeruginosa. We also demonstrate that PKCδ-depleted BMDMs and BMDMs treated with PKCδ kinase inhibitor Rottlerin fail to phosphorylate NLRC4 and activate NLRC4 inflammasome in response to P. aeruginosa infection.
PKCδ phosphorylation has also been shown to occur and be required for the NLRC4 inflammasome activity in BMDMs in response to T3SS of S. Typhimurium72. However, the identity of the kinase-mediating PKCδ phosphorylation has remained unknown for nearly a decade. We show that during infection with P. aeruginosa and in response to T3SS, Abl tyrosine kinase interacts with PKCδ and this interaction is required for PKCδ phosphorylation and for NLRC4 inflammasome activation. Abl has not been implicated in inflammasome activity or in tissue defenses against P. aeruginosa. Moreover, we show that during infection with P. aeruginosa and in response to T3SS, CrkII isoform of Crk adaptor protein interacts with Abl tyrosine kinase and this interaction is required for Abl kinase autophosphorylation (transactivation) and for the subsequent phosphorylation cascade through Abl → PKCδ → NLRC4, and ultimately for NLRC4 inflammasome assembly and activity. Crk adaptor protein has not been implicated in inflammasome activity or pro-inflammatory responses to P. aeruginosa. Of note, while Abl co-immunoprecipitated with both CrkII and PKCδ and PKCδ co-immunoprecipitated with Abl and NLRC4 in response to P. aeruginosa T3SS, CrkII and PKCδ or Abl and NLRC4 did not co-immunoprecipitate with each other, suggesting that these interactions are either transient or they are mutually exclusive as they may involve the same interaction domains in these proteins. More studies are needed to tease out the nature of these interactions.
ExoT is the only T3SS effector protein that is expressed in all T3SS-expressing P. aeruginosa clinical isolates15,105, suggesting a more fundamental role for this virulence factor in P. aeruginosa pathogenesis. In this report, we demonstrate that ExoT is the main anti-inflammatory agent for P. aeruginosa in wound and without ExoT, P. aeruginosa is substantially attenuated in its ability to colonize wound, due to heightened inflammatory responses triggered by caspase-1-dependent NLRC4 inflammasome. ExoT has not been implicated in inflammasome regulation or inflammatory responses and ExoT’s anti-inflammatory activity is perhaps the most important function for this virulence factor, adding to its growing virulence activities that make this virulence factor indispensable to P. aeruginosa pathogenesis9,10,12,52,100,102. Interestingly, we also find ExoS (but not ExoU), to contribute to P. aeruginosa fitness in wound. ExoU has been shown to significantly enhance P. aeruginosa fitness in systemic infection25. Combined, these data suggest that ExoS and ExoU enhance P. aeruginosa’s capabilities to expand its niches within the host during infection.
Imatinib treatment in cancer patients suffering from chronic myelogenous leukemia (CML), gastrointestinal stromal tumors (GISTs), and other types of cancers106,107, has been linked to increased bacterial infection, such as P. aeruginosa108,109,110,111,112, suggesting that increased vulnerability to infection in these patient cohorts could be due to dampened NLRC4 inflammasome activity.
In summary (Fig. 8), we report that P. aeruginosa T3SS triggers a rapid phosphorylation cascade involving CrkII/Abl → PKCδ → NLRC4, which leads to caspase-1 activation, culminating in the production of IL-1β and IL-18 pro-inflammatory cytokines, which in turn mobilize inflammatory leukocytes, such as neutrophils (PMNs) and macrophages (Møs), to the site of infection where they combat P. aeruginosa infection. P. aeruginosa solves this dilemma by deploying ExoT, which inhibits NLRC4 inflammasome activation by disrupting CrkII/Abl interaction, which in turn interferes with the phosphorylation cascade through the Abl/PKCδ/NLRC4 inflammasome. By functioning as an anti-inflammatory agent, ExoT enables P. aeruginosa to keep its T3SS and add other T3SS effector-virulence factors (i.e., ExoS and ExoU), which further enhance its ability to expand its niches within the host.
Insertion of T3SS in target BMDMs results in CrkII/Abl interaction which induces Abl transactivation/autophosphorylation. Activated Abl kinase then activates PKCδ kinase by phosphorylation, which in turn activates NLRC4 by phosphorylation, which then in association with ASC and pro-caspase-1 forms NLRC4 inflammasome, culminating in caspase-1 activation by proteolysis. Activated caspase-1 then processes pro-IL-1β and pro-IL-18 into mature IL-1β and IL-18, which are then secreted and in turn initiate inflammatory responses by recruiting leukocytes, such as neutrophils (PMNs) and macrophages (Møs) to the site of infection where they combat infection. P. aeruginosa ExoT dampens NLRC4 inflammasome by interfering with CrkII/Abl interaction and by blocking the phosphorylation cascade through Abl→PKCδ→NLRC4 needed for NLRC4 assembly and function.
Methods
Procedures related to animal studies
Our research complies with all relevant ethical regulations from Rush Institutional Animal Care and Use Committee. We have an approval from Rush Institutional Animal Care and Use Committee (IACUC No: 18-013) to conduct research as indicated. All procedures complied strictly with the standards for care and use of animal subjects as stated in the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Academy of Sciences, Bethesda, MD, USA). Mice were allowed to acclimate to the environment for 1 week prior to experimentation. Mice were maintained in a 12-/12-h dark/light cycle at controlled temperature (22 °C ± 1.5) and humidity conditions (40–60%) and were fed standard ad libitum diet (Teklad 2018). About 7-week-old C57BL/6 (000664), Nlrp1–/– (021301), Nlrp3–/– (021302), Cas-1–/– (032662), Cas-11–/– (024698), Pyrin+/– (Mefv+/–) (021315), and Aim2–/– (013144) mice were obtained from the Jackson Laboratories (Bar Harbor, ME). Pyrin+/– (Mefv+/–) mice were intercrossed to generate Pyrin–/– (Mefv–/–)-knockout mice. ASC–/– and Nlrc4–/– knockout mice were kind gifts from Dr. Vishva Dixit (Genentech, San Francisco, CA). All knockout mice were confirmed by genotyping. Abl flox/flox LysM Cre- and Abl flox/flox LysM Cre + were generated by crossing mice carrying floxed Abl alleles with mice carrying the LysM Cre allele97,113. All mice were backcrossed to C57BL/6 at least 6 times during generation. Femur and tibia bones of Abl flox/flox LysM Cre- and Abl flox/flox LysM Cre + were harvested as described111. (For detailed information on mice and their sources see Supplementary Table 1).
Bacteria preparation
(For a complete list of bacteria, plasmids, and their sources used in these studies, refer to Supplementary Table 1). Strains were either in PA103 or PAK genetic backgrounds and were previously described (10,13,100,114 & in Supplementary Table 1). WI-14 and WI-16 wound clinical P. aeruginosa isolates were generous gifts from Dr. Alan Hauser (Northwestern University) and were described previously15. P. aeruginosa strains were cultured in Luria–Bertani broth overnight, diluted 1:40 (volume/volume), and grown for 2 h. Bacteria were washed and resuspended in PBS.
Wounding and wound infection
Wounding was carried out as described previously47,54,57,115. A sterile biopsy punch (5-mm diameter) was used to punch through the full thickness of the back skin below the shoulder blades of mice. Bacteria were prepared as described above and 1000 bacteria (resuspended in 10 μL of saline solution) were added to each wound. Mice were housed in individual cages after wounding to prevent cross-contamination. In an experiment evaluating the role of Abl kinase in vivo, C57BL/6 mice were injected interperitoneally (i.p) with 100 mg/kg Imatinib or saline at 48, 24, and 2 h before wounding and bacterial infection. Bacteria numbers in wounds were determined as described previously47,54,69,70. Briefly, wound tissues were harvested at indicated timepoints after wounding and infection. Tissues were cut into small pieces with sterile scissors and digested with collagenase D for 40 min at 37 °C. The total number of bacteria per gram of wound tissue was then determined by serial dilution and plating on LB agar plates after normalization to tissue weight.
Bone marrow-derived macrophages (BMDMs)
BMDMs were extracted from femurs and tibias of indicated mice as described57,115. Cells were seeded in 96- or 6-well plates. Cells were primed with ultrapure LPS (Invivogen, USA) for 2–6 h and infected with P. aeruginosa strains at indicated multiplicity of infection (M.O.I) of 10–20 for 1–2 h as indicated in the figure legends. The lower M.O.I and the lower duration of infection was used for the studies involving NLRC4/ASC/caspase-1 inflammasome assessments by IF microscopy, as ExoU is highly toxic to cells116. In experiments involving the assessments of tyrosine kinases’ role in NLRC4 inflammasome, BMDMs were pretreated with indicated inhibitors: FAK inhibitor, 1,2,4,5-benzenetetraamine tetrahydrochloride at 100 μM concentration89; Src inhibitor, PP2 at 10 μM concentration90,91; Lck inhibitor, TC-S7003 at 10 μM concentration92; Fyn inhibitor, AZD0530 at 10 μM concentration93; Lyn inhibitor, Lyn peptide inhibitor at 20 μM concentration94; Abl inhibitor, Imatinib at 30 μM95; or PKCδ inhibitor, Rottlerin at 5 µM, for 90 min before bacterial infection.
Cytokine analyses
Tissue homogenates and cell culture supernatants were assessed for the pro-inflammatory cytokines IL-1β and IL-18 by ELISA (Invitrogen, USA) according to the manufacturer’s instructions, as we described54,57,60.
Histopathological evaluation
Leukocytes’ infiltration in wounds was assessed by immunohistochemical (IHC) analyses using H&E, as described previously47,54,57,58,117. Neutrophil activation in wounds was assessed by IHC analysis using MPO staining (anti-Myeloperoxidase antibody) (Abcam Cat. No. ab9535, 1:200)54,58,60,70. The histological data were obtained from N ≥ 5 mice/group and ≥7 random fields/wound/mouse.
Western blot analyses
Western blot analysis were performed as described previously13,52,114,118. Briefly, the supernatants of treated and/or infected BMDMs were subjected to trichloroacetic acid protein precipitation. The precipitates were denatured in Laemmli buffer and analyzed by Western blot for activated caspase-1 and IL-1β, using their respective antibodies. Cell lysates were immunoblotted for pro-caspase-1 and pro-IL-1β or immunoblotted for indicated proteins (phosphorylated and unphosphorylated forms) using their respective antibodies.
Antibodies
Anti-caspase-1 (P20) (AdipoGen Cat. No. AG20B0042C100, 1:1000), anti-ASC pab (AL177) (AdipoGen Cat. No. AG25B0006C100, 1:1000), Crk Mouse Monoclonal Antibody Clone: 22 (BD Cat. No. 610035, 1:1000), NLRC4 (Abcam Cat. No. ab201792 and Millipore Sigma Cat. No. 06-112-5MI, 1:200), Phospho-NLRC4 (SER-533) PAB (ECM Biosciences Cat. No. NP5411, 1:50), Phospho-NLRC4 (Ser533) Mouse anti-Human, Mouse, Clone: 4B7B7 (Invitrogen Cat. No. PIMA531846, 1:50), Polyclonal Anti-PKCδ (Cell Signaling Technology Cat. No. 2058, 1:1000), Phospho-PKCδ (Tyr311) Antibody (Cell Signaling Technology Cat. No. 2055, 1:500), and Mouse IL-1β Antibody (R&D systems Cat. No. AF-401-NA, 1:1000). Mouse (MOPC-21) mAb IgG1 Isotype Control (Cell Signaling Technology, Cat. No. 4097, 1:1000) NLRC4 Polyclonal Antibody Invitrogen (Cat. No. PA5-72908, 1:100), Anti- c-Abl (24-11) (Santa Cruz Biotechnology Cat. No. sc-23, 1:100), donkey anti-goat IGG Secondary Antibody (HRP) (Novus Biologicals Cat. No. NBP1-74815, 1:1000), β-Actin (13E5) Rabbit mAb (Cell Signaling Technology, Cat. No. 4970, 1:2000) Anti-Myeloperoxidase antibody (Abcam Cat. No. ab9535, 1:1000), Anti-ABL1 (phosphor-Y245) (Abcam Cat. No. ab193223, 1:100), anti-rabbit IGG, HRP-linked Antibody (Cell Signaling Technology, Cat. No. 7074 S, 1:5000), Anti-mouse IgG, HRP-linked Antibody (Cell Signaling Technology, Cat. No. 7076, 1:5000), and GAPDH Antibody Rabbit Polyclonal (Proteintech Cat. No. 1094-I-AP, 1:5000).
Immunoprecipitation
BMDMs were infected with indicated P. aeruginosa strains at M.O.I of 20 for 2 h. Whole-cell lysates were immunoprecipitated with antibodies against NLRC4 (NLRC4 Polyclonal Antibody (Invitrogen Cat. No. PA5-72908 and Abcam Cat. No. ab201792, 1:50)), PKCδ (Cell Signaling Technology, Cat. No. 2058, 1:50) and Crk (BD Cat. No. 610035, 1:50), and Abl (Santa Cruz Biotechnology Cat. No. sc-23, 1:50) according to the manufacturer’s instructions (Direct and Classic ip kits, Pierce). Immunoprecipitated lysates were immunoblotted for indicated proteins as discussed earlier.
Protein depletion by siRNA transfection
Protein depletion by siRNA was performed as described9,82,119. Briefly, BMDMs were transfected with Crk siRNA, PKCδ siRNA, and Abl siRNA (final concentration, 100 nM) using a lipofectamine RNAimax reagent (Invitrogen, USA) according to the manufacturer’s instructions for 48 h, 48 h, and 24 h, respectively. After transfection, cells were washed with PBS to remove transfection regents and resuspended in fresh media for the indicated experiments. Cells were primed with ultrapure LPS (200 ng/mL) for 2 h. They were then infected with indicated P. aeruginosa strains at M.O.I. of 20 for 2 h. Whole-cell lysates were assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting, and the corresponding supernatants were assessed for IL-1β and IL-18 cytokines by ELISA as discussed earlier.
Nlrc4 −/− cells’ reconstitution
Nlrc4−/− cells (obtained from Genentech) were reconstituted with wild-type NLRC4 or nonphosphorylatable NLRC4 mutant (NLRC4/S533A) vectors, as described72 and according to the manufacturer’s instructions (Cell Biolabs). Reconstituted cells were infected with P. aeruginosa (M.O.I. 20) for 2 h. Whole-cell lysates were immunoprecipitated with antibodies against Flag (Millipore Sigma, Cat. No. F1804, 1:50) and assessed for indicated proteins (phosphorylated and unphosphorylated forms) by Western blotting. The culture supernatants were subjected to trichloroacetic acid protein precipitation. The precipitates were denatured in Laemmli buffer and analyzed by Western blot for activated caspase-1. The culture supernatants were assessed for IL-1β and IL-18 cytokines by ELISA as discussed earlier.
Transient transfection was performed as previously described13,52,102. Briefly, BMDMs were pretreated with a pan-caspase inhibitor (Z-VAD) at 60 µM final concentration 1 h prior to transfection with indicated plasmids (ExoT-GFP, ExoT (G-A+)–GFP, ExoT (G+A−)–GFP, and GFP) to prevent ExoT/GAP or ExoT/ADPRT-induced apoptosis10,100. About 24 h after transfection, cells were either treated with PBS (uninfected) or infected with indicated P. aeruginosa strains for 2 h. Whole-cell lysates were immunoprecipitated with antibodies against GFP (Abcam, Cat. No. AB5450, 1:50) and immunoblotted for indicated proteins.
Fluorescence microscopy
Immunofluorescent microscopy was performed as described12,13,82,114. Briefly, BMDMs were seeded in coated coverslips in 24-well plates. After LPS priming and bacterial infection, coverslips were fixed with 4% PFA for 20 mins, and permeabilized using 0.1% Triton X-100 and blocked for 60 mins at room temperature. Coverslips were stained with antibodies against ASC (AdipoGenCat. No. AG25B0006C100, 1:200), Caspase-1 (AdipoGen Cat. No. AG20B0042C100, 1:200), and p-NLRC4 (Invitrogen Cat. No. PIMA531846, 1:100) at 1:100 dilution overnight at 4 °C. After washing with 1X PBS, coverslips were then incubated with goat anti-mouse IgG (H + L) secondary antibody, Alexa Fluor 488 (Invitrogen Cat. No. A-11001, 1:500), goat anti-rabbit IgG (H + L) secondary antibody, and Texas Red (Invitrogen Cat. No. T-6391, 1:500), at 1:500 dilution for 1 h at room temperature in the dark. Coverslips were washed three times with 1 X PBS, mounted with DAPI nuclear dye, and visualized under fluorescence microscope. These studies were done in triplicates, and for each replicate, ≥7 randomly selected fields were counted, and data were presented as the mean ± SEM.
Data collection
Imaging data were collected on Axiovert Z1 microscope (ZEISS), using ZEISS ZEN blue edition software.
Data analyses
We used GraphPad Prism version 8 (GraphPad software), Excel (Microsoft Office, version, Professional Plus 2016), Image J (open source, version, 1.52a), and Illustrator 2021 (Adobe, version, 25.4.3), to analyze the data in this study.
Sample size
Sample size was determined based on our previous experience12,54,57,117,120 and was large enough to detect differences between samples with biological significance.
Quantification and statistical analysis
Statistical analyses between two groups were performed by two-sided unpaired Student’s t-test and statistical analyses of more than two groups were performed by one-way analysis of variance (ANOVA) with additional post hoc testing, using the GraphPad Prism software, version 8. Data are presented as mean ± SEM or mean ± SD. P-values less than or equal to 0.05 were taken as significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
A “Reporting Summary” for this article is available as Supplementary Information file. The main data supporting the findings of this study are available within the article and its Supplementary Figures. The source data underlying all figures and Supplementary Figures are provided as a Source Data file. Source data are provided with this paper.
References
Koch, C. Early infection and progression of cystic fibrosis lung disease. Pediatr. Pulmonol. 34, 232–236 (2002).
Engel, J. N. in Severe Infections Caused by Pseudomonas aeruginosa (eds Hauser, A. & Rello, J.) 201–230 (Kluwer Academic/Plenum Press, 2003).
Mahajan-Miklos, S., Tan, M. W., Rahme, L. G. & Ausubel, F. M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96, 47–56 (1999).
Kamath, S., Kapatral, V. & Chakrabarty, A. M. Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol. Microbiol 30, 933–941 (1998).
Matsumoto, K. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 385, 1007–1016 (2004).
Feldman, M. et al. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 66, 43–51 (1998).
Hauser, A. R. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665 (2009).
Engel, J. & Balachandran, P. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 12, 61–66 (2009).
Shafikhani, S. H. & Engel, J. Pseudomonas aeruginosa type III-secreted toxin ExoT inhibits host-cell division by targeting cytokinesis at multiple steps. Proc. Natl Acad. Sci. USA 103, 15605–15610 (2006).
Wood, S. J., Goldufsky, J. & Shafikhani, S. H. Pseudomonas aeruginosa ExoT Induces Atypical Anoikis Apoptosis in Target Host Cells by Transforming Crk Adaptor Protein into a Cytotoxin. PLoS Pathog. 11, e1004934 (2015).
Wood, S. J., Goldufsky, J. W., Bello, D., Masood, S. & Shafikhani, S. H. Pseudomonas aeruginosa ExoT induces mitochondrial apoptosis in target host cells in a manner that depends on its GAP domain activity. J. Biol. Chem., https://doi.org/10.1074/jbc.M115.689950 (2015).
Gupta, K. H. et al. Apoptosis and compensatory proliferation signaling are coupled by CrkI-containing microvesicles. Dev. Cell 41, 674–684 e675 (2017).
Kaminski, A. et al. Pseudomonas aeruginosa ExoS induces intrinsic apoptosis in target host cells in a manner that is dependent on its GAP domain activity. Sci. Rep. 8, 14047 (2018).
Stavrinides, J., McCann, H. C. & Guttman, D. S. Host-pathogen interplay and the evolution of bacterial effectors. Cell Microbiol. 10, 285–292 (2008).
Feltman, H. et al. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147, 2659–2669 (2001).
Wolfgang, M. C. et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 100, 8484–8489 (2003).
Coburn, B., Sekirov, I. & Finlay, B. Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549 (2007).
Franchi, L. et al. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur. J. Immunol. 37, 3030–3039 (2007).
Miao, E. A., Ernst, R. K., Dors, M., Mao, D. P. & Aderem, A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc. Natl Acad. Sci. USA 105, 2562–2567 (2008).
Balakrishnan, A., Karki, R., Berwin, B., Yamamoto, M. & Kanneganti, T.-D. Guanylate binding proteins facilitate caspase-11-dependent pyroptosis in response to type 3 secretion system-negative Pseudomonas aeruginosa. Cell Death Discov. 4, 1–12 (2018).
Bitto, N. J. et al. Membrane vesicles from Pseudomonas aeruginosa activate the noncanonical inflammasome through caspase‐5 in human monocytes. Immunol. Cell Biol. 96, 1120–1130 (2018).
Karmakar, M., Sun, Y., Hise, A. G., Rietsch, A. & Pearlman, E. Cutting edge: IL-1β processing during Pseudomonas aeruginosa infection is mediated by neutrophil serine proteases and is independent of NLRC4 and caspase-1. J. Immunol. 189, 4231–4235 (2012).
Gugliandolo, E. et al. Involvement of TLR4 and PPAR-α receptors in host response and NLRP3 inflammasome activation, against pulmonary infection with Pseudomonas aeruginosa. Shock 51, 221–227 (2019).
Lin, C. K. & Kazmierczak, B. I. Inflammation: a double-edged sword in the response to Pseudomonas aeruginosa infection. J. Innate Immun. 9, 250–261 (2017).
Sutterwala, F. S. et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204, 3235–3245 (2007).
Rimessi, A. et al. Mitochondrial Ca2+-dependent NLRP3 activation exacerbates the Pseudomonas aeruginosa-driven inflammatory response in cystic fibrosis. Nat. Commun. 6, 1–16 (2015).
Deng, Q. et al. Pseudomonas aeruginosa triggers macrophage autophagy to escape intracellular killing by activation of the NLRP3 inflammasome. Infect. Immun. 84, 56–66 (2016).
Iatridou, K., Mandalidis, D., Chronopoulos, E., Vagenas, G. & Athanasopoulos, S. Static and dynamic body balance following provocation of the visual and vestibular systems in females with and without joint hypermobility syndrome. J. Bodyw. Mov. Ther. 18, 159–164 (2014).
Jabir, M. S. et al. Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and β-interferon production during Pseudomonas aeruginosa infection. Cell Host Microbe 15, 214–227 (2014).
Iannitti, R. G. et al. IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis. Nat. Commun. 7, 1–16 (2016).
Faure, E. et al. Pseudomonas aeruginosa type-3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. Am. J. Respir. Crit. Care Med. 189, 799–811 (2014).
Galle, M. et al. The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1beta maturation. J. Cell Mol. Med. 12, 1767–1776 (2008).
de Lima, C. D. M. et al. ExoU activates NF-κB and increases IL-8/KC secretion during Pseudomonas aeruginosa infection. PloS ONE 7, e41772 (2012).
Diaz Ferrer, J. et al. [Virological breakthrough in treating hepatitis B with entecavir: it means resistance?]. Rev. Gastroenterol. Peru. Organo of. Soc. Gastroenterol. Peru. 32, 400–404 (2012).
Saliba, A. M. et al. Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cell Microbiol. 7, 1811–1822 (2005).
Epelman, S., Bruno, T. F., Neely, G. G., Woods, D. E. & Mody, C. H. Pseudomonas aeruginosa exoenzyme S induces transcriptional expression of proinflammatory cytokines and chemokines. Infect. Immun. 68, 4811–4814 (2000).
Epelman, S. et al. Distinct fates of monocytes and T cells directly activated by Pseudomonas aeruginosa exoenzyme S. J. Leukoc. Biol. 71, 458–468 (2002).
Epelman, S. et al. Different domains of Pseudomonas aeruginosa Exoenzyme S activate distinct TLRs. J. Immunol. 173, 2031–2040 (2004).
Yamaguchi, T. & Yamada, H. Role of mechanical injury on airway surface in the pathogenesis of Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 144, 1147–1152 (1991).
Zahm, J. M., Chevillard, M. & Puchelle, E. Wound repair of human surface respiratory epithelium. Am. J. Respir. Cell Mol. Biol. 5, 242–248 (1991).
Tsang, K. W. T. et al. Interaction of Pseudomonas aeruginosa with human respiratory mucosa in vitro. Eur. Respir. J. 7, 1746–1753 (1994).
de Bentzmann, S., Plotkowski, C. & Puchelle, E. Receptors in the Pseudomonas aeruginosa adherence to injured and repairing airway epithelium. Am. J. Respir. Crit. Care Med. 154, S155–S162 (1996).
Madsen, S. M., Westh, H., Danielsen, L. & Rosdahl, V. T. Bacterial colonization and healing of venous leg ulcers. APMIS 104, 895–899 (1996).
Halbert, A. R., Stacey, M. C., Rohr, J. B. & Jopp-McKay, A. The effect of bacterial colonization on venous ulcer healing. Australas. J. Dermatol. 33, 75–80 (1992).
Gjodsbol, K. et al. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int. Wound J. 3, 225–231 (2006).
Winstanley, C. et al. Genotypic and phenotypic characteristics of Pseudomonas aeruginosa isolates associated with ulcerative keratitis. J. Med. Microbiol. 54, 519–526 (2005).
Goldufsky, J. et al. Pseudomonas aeruginosa uses T3SS to inhibit diabetic wound healing. Wound Repair Regen. 23, 557–564 (2015).
Garrity-Ryan, L. et al. The ADP ribosyltransferase domain of Pseudomonas aeruginosa ExoT contributes to its biological activities. Infect. Immun. 72, 546–558 (2004).
Muller, M., Li, Z. & Maitz, P. K. Pseudomonas pyocyanin inhibits wound repair by inducing premature cellular senescence: role for p38 mitogen-activated protein kinase. Burns: J. Int. Soc. Burn Injuries 35, 500–508 (2009).
Heggers, J. P. et al. Pseudomonas aeruginosa Exotoxin A: Its Role in Retardation of Wound Healing The 1992 Lindberg Award. J. Burn Care Rehabil. 13, 512–518 (1992).
Watters, C. et al. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med. Microbiol. Immunol. 202, 131–141 (2013).
Mohamed, M. F. et al. Pseudomonas aeruginosa ExoT induces G1 cell cycle arrest in melanoma cells. Cell Microbiol. 23, e13339 (2021).
Gellatly, S. L. & Hancock, R. E. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173 (2013).
Roy, R. et al. Overriding impaired FPR chemotaxis signaling in diabetic neutrophil stimulates infection control in murine diabetic wound. eLife, https://doi.org/10.7554/eLife.72071 (2022).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Van de Veerdonk, F. L., Netea, M. G., Dinarello, C. A. & Joosten, L. A. Inflammasome activation and IL-1β and IL-18 processing during infection. Trends Immunol. 32, 110–116 (2011).
Roy, R. et al. IL-10 Dysregulation Underlies Chemokine Insufficiency, Delayed Macrophage Response, and Impaired Healing in Diabetic Wound. J. Invest. Dermatol. 142, 692–704.e14 (2022).
Kroin, J. S. et al. Perioperative high inspired oxygen fraction therapy reduces surgical site infection with Pseudomonas aeruginosa in rats. J. Med. Microbiol. 65, 738–744 (2016).
Jeong, W. et al. Scar Prevention and Enhanced Wound Healing Induced by Polydeoxyribonucleotide in a Rat Incisional Wound-Healing Model. Int. J. Mol. Sci., https://doi.org/10.3390/ijms18081698 (2017).
Mahmud, F. et al. Therapeutic evaluation of immunomodulators in reducing surgical wound infection. FASEB J. 36, e22090 (2022).
Klebanoff, S. J. Myeloperoxidase: friend and foe. J. Leukoc. Biol. 77, 598–625 (2005).
Kayagaki, N. et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117 (2011).
Kayagaki, N. et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015).
Cohen, T. S. & Prince, A. S. Activation of inflammasome signaling mediates pathology of acute P. aeruginosa pneumonia. J. Clin. Investig. 123, 1630–1637 (2013).
Broz, P., von Moltke, J., Jones, J. W., Vance, R. E. & Monack, D. M. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483 (2010).
Sharma, D. & Kanneganti, T.-D. The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J. Cell Biol. 213, 617–629 (2016).
Malik, A. & Kanneganti, T. D. Inflammasome activation and assembly at a glance. J. Cell Sci. 130, 3955–3963 (2017).
Miao, E. A. et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107, 3076–3080 (2010).
Kroin, J. S. et al. Short-term glycemic control is effective in reducing surgical site infection in diabetic rats. Anesth. Analg., https://doi.org/10.1213/ANE.0000000000000650 (2015).
Kroin, J. S. et al. Perioperative high inspired oxygen fraction therapy reduces surgical site infection with Pseudomonas aeruginosa in rats. J. Med. Microbiol., https://doi.org/10.1099/jmm.0.000295 (2016).
Mariathasan, S. et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004).
Qu, Y. et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490, 539–542 (2012).
Matusiak, M. et al. Flagellin-induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proc. Natl Acad. Sci. USA 112, 1541–1546 (2015).
Liu, W. et al. LRRK2 promotes the activation of NLRC4 inflammasome during Salmonella Typhimurium infection. J. Exp. Med. 214, 3051–3066 (2017).
Suzuki, S. et al. Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcdelta. PLoS Pathog. 10, e1003926 (2014).
Tenthorey, J. L. et al. NLRC4 inflammasome activation is NLRP3-and phosphorylation-independent during infection and does not protect from melanoma. J. Exp. Med. 217, e20191736 (2020).
Sataranatarajan, K., Lee, M. J., Mariappan, M. M. & Feliers, D. PKCdelta regulates the stimulation of vascular endothelial factor mRNA translation by angiotensin II through hnRNP K. Cell. Signal. 20, 969–977 (2008).
Krall, R., Schmidt, G., Aktories, K. & Barbieri, J. T. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect. Immun. 68, 6066–6068 (2000).
Sun, J. & Barbieri, J. T. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J. Biol. Chem. 278, 32794–32800 (2003).
Mayer, B. J., Hamaguchi, M. & Hanafusa, H. A novel viral oncogene with structural similarity to phospholipase C. Nature 332, 272–275 (1988).
Mayer, B. J. & Hanafusa, H. Mutagenic analysis of the v-Crk oncogene: requirement for SH2 and SH3 domains and correlation between increased cellular phosphotyrosine and transformation. J. Virol. 64, 3581–3589 (1990).
Wood, S., Sivaramakrishnan, G., Engel, J. & Shafikhani, S. H. Cell migration regulates the kinetics of cytokinesis. Cell Cycle 10, 648–654 (2011).
Iwahara, T., Akagi, T., Fujitsuka, Y. & Hanafusa, H. CrkII regulates focal adhesion kinase activation by making a complex with Crk-associated substrate, p130Cas. Proc. Natl Acad. Sci. USA 101, 17693–17698 (2004).
Gelkop, S., Babichev, Y., Kalifa, R., Tamir, A. & Isakov, N. Involvement of crk adapter proteins in regulation of lymphoid cell functions. Immunol. Res. 28, 79–91 (2003).
Park, T.-J. & Curran, T. Crk and Crk-like play essential overlapping roles downstream of disabled-1 in the Reelin pathway. J. Neurosci. 28, 13551–13562 (2008).
Schmoker, A. M. et al. Dynamic multi-site phosphorylation by Fyn and Abl drives the interaction between CRKL and the novel scaffolding receptors DCBLD1 and DCBLD2. Biochem. J. 474, 3963–3984 (2017).
Braiman, A. & Isakov, N. The Role of Crk Adaptor Proteins in T-Cell Adhesion and Migration. Front Immunol. 6, 509 (2015).
He, Y. et al. The non-receptor tyrosine kinase Lyn controls neutrophil adhesion by recruiting the CrkL-C3G complex and activating Rap1 at the leading edge. J. Cell Sci. 124, 2153–2164 (2011).
Golubovskaya, V. M. et al. A small molecule inhibitor, 1,2,4,5-benzenetetraamine tetrahydrochloride, targeting the y397 site of focal adhesion kinase decreases tumor growth. J. Med. Chem. 51, 7405–7416 (2008).
Brandvold, K. R., Steffey, M. E., Fox, C. C. & Soellner, M. B. Development of a highly selective c-Src kinase inhibitor. ACS Chem. Biol. 7, 1393–1398 (2012).
Malin, D. et al. ERK-regulated αB-crystallin induction by matrix detachment inhibits anoikis and promotes lung metastasis in vivo. Oncogene 34, 5626–5634 (2015).
Martin, M. W. et al. Structure-based design of novel 2-amino-6-phenyl-pyrimido[5’,4’:5,6]pyrimido[1,2-a]benzimidazol-5(6H)-ones as potent and orally active inhibitors of lymphocyte specific kinase (Lck): synthesis, SAR, and in vivo anti-inflammatory activity. J. Med. Chem. 51, 1637–1648 (2008).
Du, G. et al. Targeting Src family kinase member Fyn by Saracatinib attenuated liver fibrosis in vitro and in vivo. Cell Death Dis. 11, 118 (2020).
Adachi, T., Stafford, S., Sur, S. & Alam, R. A novel Lyn-binding peptide inhibitor blocks eosinophil differentiation, survival, and airway eosinophilic inflammation. J. Immunol. 163, 939–946 (1999).
Lin, Y.-L., Meng, Y., Jiang, W. & Roux, B. Explaining why Gleevec is a specific and potent inhibitor of Abl kinase. Proc. Natl Acad. Sci. USA 110, 1664–1669 (2013).
Yang, F.-C. et al. Nf1+/− mast cells induce neurofibroma like phenotypes through secreted TGF-β signaling. Hum. Mol. Genet. 15, 2421–2437 (2006).
Wetzel, D. M., Rhodes, E. L., Li, S., McMahon-Pratt, D. & Koleske, A. J. The Src kinases Hck, Fgr and Lyn activate Arg to facilitate IgG-mediated phagocytosis and Leishmania infection. J. Cell Sci. 129, 3130–3143 (2016).
Shishido, T. et al. Crk family adaptor proteins trans‐activate c‐Abl kinase. Genes Cells 6, 431–440 (2001).
Reichman, C. et al. Transactivation of Abl by the Crk II adapter protein requires a PNAY sequence in the Crk C-terminal SH3 domain. Oncogene 24, 8187–8199 (2005).
Wood, S. J., Goldufsky, J. W., Bello, D., Masood, S. & Shafikhani, S. H. Pseudomonas aeruginosa ExoT induces mitochondrial apoptosis in target host cells in a manner that depends on its GTPase-activating protein (GAP) domain activity. J. Biol. Chem. 290, 29063–29073 (2015).
Goldufsky, J. et al. Pseudomonas aeruginosa exotoxin T induces potent cytotoxicity against a variety of murine and human cancer cell lines. J. Med. Microbiol. 64, 164–173 (2015).
Shafikhani, S. H., Morales, C. & Engel, J. The Pseudomonas aeruginosa type III secreted toxin ExoT is necessary and sufficient to induce apoptosis in epithelial cells. Cell. Microbiol. 10, 994–1007 (2008).
Koleske, A. J. et al. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21, 1259–1272 (1998).
Chislock, E. M., Ring, C. & Pendergast, A. M. Abl kinases are required for vascular function, Tie2 expression, and angiopoietin-1–mediated survival. Proc. Natl Acad. Sci. USA 110, 12432–12437 (2013).
Feltman, H., Khan, S., Jain, M., Peterson, L. & Hauser, A. Type III secretion genotypes of clinical and environmental Pseudmonas aeruginosa isolates. ASM Abstracts, D22 (2001).
Biondi, A. et al. Imatinib treatment of paediatric Philadelphia chromosome-positive acute lymphoblastic leukaemia (EsPhALL2010): a prospective, intergroup, open-label, single-arm clinical trial. Lancet Haematol. 5, e641–e652 (2018).
Laurent, M. et al. Adjuvant therapy with imatinib in gastrointestinal stromal tumors (GISTs)—review and perspectives. Transl. Gastroenterol. Hepatol. 4, 24 (2019).
Suzuki, K. & Sekine, T. [A patient with acute Philadelphia-chromosome-positive mixed phenotype leukemia developing ecthyma gangrenosum while undergoing combined imatinib mesylate chemotherapy. Kansenshogaku zasshi. J. Jpn. Assoc. Infect. Dis. 83, 669–672 (2009).
Abugroun, A., Chaudhary, A. & Rodriguez, G. Fatal outcome of imatinib in a patient with Idiopathic Hypereosinophilic Syndrome. Case Rep. Oncol. Med. https://doi.org/10.1155/2018/6291614 (2018).
Thia, T., Tan, H., Chuah, T., Chow, W. & Lui, H. Imatinib mesylate-related fatal acute hepatic failure in a patient with chronic myeloid leukaemia and chronic hepatitis B infection. Singap. Med. J. 49, e86 (2008).
Lakhani, S., Davidson, L., Priebat, D. A. & Sherker, A. H. Reactivation of chronic hepatitis B infection related to imatinib mesylate therapy. Hepatol. Int. 2, 498–499 (2008).
Mattiuzzi, G. N. et al. Development of Varicella-Zoster virus infection in patients with chronic myelogenous leukemia treated with imatinib mesylate. Clin. Cancer Res. 9, 976–980 (2003).
Wetzel, D. M., McMahon-Pratt, D. & Koleske, A. J. The Abl and Arg kinases mediate distinct modes of phagocytosis and are required for maximal Leishmania infection. Mol. Cell. Biol. 32, 3176–3186 (2012).
Mahmood, F. et al. A novel human antimicrobial factor targets Pseudomonas aeruginosa through its type III secretion system. J. Med. Microbiol. 62, 531–539 (2013).
Wood, S. et al. Pro-inflammatory chemokine CCL2 (MCP-1) promotes healing in diabetic wounds by restoring the macrophage response. PloS ONE 9, e91574 (2014).
Finck-Barbancon, V. & Frank, D. W. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J. Bacteriol. 183, 4330–4344 (2001).
Hamilton, J. L., Mohamed, M. F., Witt, B. R., Wimmer, M. A. & Shafikhani, S. H. Therapeutic assessment of N-formyl-methionyl-leucyl-phenylalanine (fMLP) in reducing periprosthetic joint infection. Eur. Cell Mater. 41, 122–138 (2021).
Wood, S. et al. Chronic alcohol exposure renders epithelial cells vulnerable to bacterial infection. PloS ONE 8, e54646 (2013).
Shafikhani, S. H., Mostov, K. & Engel, J. Focal adhesion components are essential for mammalian cell cytokinesis. Cell Cycle 7, 2868–2876 (2008).
Kroin, J. S. et al. Local vancomycin effectively reduces surgical site infection at implant site in rodents. Regional Anesthesia Pain. Med. 43, 795–804 (2018).
Acknowledgements
We would like to thank Dr. Vishva Dixit and Genentech for their generous gifts of ASC−/− and Nlrc4−/− knockout mice and for NLRC4–/– BMDM and the expression vectors. We also would like to thank Dr. Alan Hauser (Northwestern University) for providing P. aeruginosa wound clinical isolates, and the rest of Shafikhani lab members for their valued opinions on these studies. This work was supported by the National Institutes of Health (NIH) grants RO1DK107713 and R21AI110685-01 (both to S.H.S.), and R01AI146349 and Welch Foundation I-2086 (to D.M.W.).
Author information
Authors and Affiliations
Contributions
S. H. S. conceived and coordinated all aspects of the studies and wrote the paper. M.F.M. conceived, conducted, and contributed to all the figures in this paper and wrote the paper. K.G. contributed to the data presented in Figs. 1–2 and Supplementary Figs 1–3. J.G. contributed to experiments involving leukocytes and neutrophil infiltration by IHC and to Fig. 8. RR contributed to IHC experiments. L.T.C and D.M.W. contributed to Fig. 6. T.K. and J.R., contributed to research design, data analyses, and reagents. All authors had the opportunity to review and edit the paper. All authors approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alice Prince and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohamed, M.F., Gupta, K., Goldufsky, J.W. et al. CrkII/Abl phosphorylation cascade is critical for NLRC4 inflammasome activity and is blocked by Pseudomonas aeruginosa ExoT. Nat Commun 13, 1295 (2022). https://doi.org/10.1038/s41467-022-28967-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-28967-5
This article is cited by
-
The role of inflammasomes in human diseases and their potential as therapeutic targets
Signal Transduction and Targeted Therapy (2024)
-
Recent advances in therapeutic targets identification and development of treatment strategies towards Pseudomonas aeruginosa infections
BMC Microbiology (2023)
-
NLRP3 selectively drives IL-1β secretion by Pseudomonas aeruginosa infected neutrophils and regulates corneal disease severity
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.