Abstract
Organic elements make up 99% of an organism but without the remaining inorganic bioessential elements, termed the metallome, no life could be possible. The metallome is involved in all aspects of life, including charge balance and electrolytic activity, structure and conformation, signaling, acid-base buffering, electron and chemical group transfer, redox catalysis energy storage and biomineralization. Here, we report the evolution with age of the metallome and copper and zinc isotope compositions in five mouse organs. The aging metallome shows a conserved and reproducible fingerprint. By analyzing the metallome in tandem with the phenome, metabolome and proteome, we show networks of interactions that are organ-specific, age-dependent, isotopically-typified and that are associated with a wealth of clinical and molecular traits. We report that the copper isotope composition in liver is age-dependent, extending the existence of aging isotopic clocks beyond bulk organic elements. Furthermore, iron concentration and copper isotope composition relate to predictors of metabolic health, such as body fat percentage and maximum running capacity at the physiological level, and adipogenesis and OXPHOS at the biochemical level. Our results shed light on the metallome as an overlooked omic layer and open perspectives for potentially modulating cellular processes using careful and selective metallome manipulation.
Similar content being viewed by others
Introduction
Knowledge of the metallome – briefly, the set of inorganic elements in an organism, (for a complete definition see Lobinski et al.1) and its interactions with other omic layers is elemental for understanding how physiological perturbations cause or result from alterations of metal concentrations and stable isotope compositions. Isotopic fractionation, the partitioning of stable isotopes due to slight differences in mass-dependent quantum zero-point energies2,3, is an emerging means for characterizing the involvement of the metallome in biological processes4,5,6. So far, interactions of the metallome with other omic layers have been mostly studied in plants to optimize crop production7,8,9. Scarcer studies in yeast10 and cultured human cells11 reveal a wealth of metallome-dependent biological traits. The present study is the first comprehensive analysis of the interactions between the metallome and other omic layers (phenome, metabolome, and proteome) in organs of mice as a function of age.
Results and discussion
The metallome and isotope compositions fingerprints are organ-specific
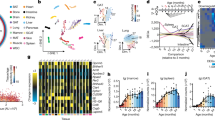
We measured the metallome (K, Mg, Na, P, S, Ca, Fe, Cu, Rb, Zn, Se, Co, Mo, and Cd) and the Cu and Zn stable isotope compositions (denoted hereafter δ65Cu and δ66Zn, respectively) in five organs (brain, heart, kidney, liver, and muscle, Supplementary data 1 and table S1) of 49 C57BL/6 male mice aged 6-, 16-, and 24-months old (mo) (Fig. 1a). Principal Component Analysis (PCA) shows organ-specific metallomic and isotopic signatures (Fig. 1b) as described before12,13,14. The first principal component is driven oppositely by protein-bound transition metals (Fe, Zn, Se, Mn, Co, and Cu) and free alkali and alkaline earth metals (Ca, K, and Mg) (Fig. 1b). The δ65Cu and δ66Zn values are also organ-specific, suggesting that isotope fractionation depends on fine, organ-specific processes (Fig. 1c, d). The δ66Zn value in heart, brain, muscle and liver follows a linear relationship with Zn concentration, a relationship that is known to indicate identical Zn routing in these organs15, obviously distinct in kidneys where glomerular filtration probably drives isotopic fractionation through a distillation process16 (Fig. 1c). Copper isotope fractionation is more intense than that of Zn. Cu exhibits a unique concentration vs isotopic composition signature for each organ, depending on the occurrence of its oxidation state, Cu(II) compounds being isotopically heavier than Cu(I) compounds17 (Fig. 1d).
a Study design and overlapping samples between omic layers. Icons are designed by www.freepik.com, and assembled by VB on Adobe Illustrator. b Principal component analysis of the metal concentration and isotope composition of each sample. Clear organ signatures are visible, with small shifts with aging. c, d Relation between isotope composition and concentrations of Cu (c) and Zn (d). Error bars indicate standard deviation. The dashed line indicates a linear relationship between δ66Zn and Zn between the liver, brain, muscle, and heart, indicative of identical Zn routing in these organs. e Volcano plots of the effect of aging on metal concentrations. Triangles: 16 vs 6 m.o., circles 16 vs 24 m.o., squares: 24 vs 6 m.o. The dotted line indicates the threshold of Adj.P < 0.05, FDR-corrected (limma) moderated t-test, two-sided. f Boxplot of the most age-dependent metal concentrations and isotope compositions. The lower and upper hinges correspond to the first and third quartiles, and center line is the median. The whiskers extend from the hinge to the largest value no further than 1.5 * inter-quartile range. ***Adj.P < 0.001, **Adj.P < 0.01, * Adj.P < 0.05, FDR-corrected (limma) moderated t-test, two-sided. g Network of Spearman Rho correlations between metals. Only correlated metals with **Adj.P < 0.001, **Adj.P < 0.01, * Adj.P < 0.05, (limma) moderated t-test, with age as a covariate, are pictured. The correlation coefficient and directionality of the correlation are pictured through line thickness and color, respectively. Source data are provided as a Source Data file.
We further explored the metallome evolution during aging. Major elements, i.e. electrolytes (K, Mg, and Na) or molecular constituents (S and P), show a remarkable homogeneity among organs and stability over time (Fig. S1A). Minor elements (Ca, Cu, Fe, Rb, Zn), with countless biological roles18, and especially ultra-trace elements – Se for selenoproteins, Co for vitamin B12, Mo for molybdoenzymes, and Cd with unknown biological role18 – exhibit a more heterogeneous distribution in the body than major elements and vary with age (Fig. S1B–S1C). Such ultra-trace elements are mainly present in the liver and to a lesser extent in the kidney and brain. The gastrocnemius striated muscle and the myocardial muscle are depleted in ultra-trace elements, probably reflecting their limited involvement in specific biochemical synthesis (Fig. S1C).
The metallome and isotope compositions fingerprints are age-dependent
Of all the organs, metal concentrations in the brain seem to vary the most with age (Fig. 1e, Rb, Fe, Ca, Cu, and Co at p-value < 0.05). We find an age-dependent accumulation of Fe and Cu in a healthy brain (Fig. 1f), which complements previous observations19,20,21,22 in the mouse. As high levels of transition metals such as Fe, Cu, and Zn are known to be associated with amyloid-β plaques and α-synuclein accumulation in neurodegenerative diseases23,24, all these results may suggest that metal accumulation precedes the formation of the protein aggregates. The accumulation of Cd in kidney (Fig. 1f) may be illustrative of chronic toxicity25. Rubidium, which is the K minor element analog26, decreases between 6 m.o. and 24 m.o. in brain, muscle, and kidney (Figs. 1f and S2), suggesting unprecedented evidence of disrupted K metabolism in old mice. These results confirm the value of the metallome as a biomarker for aging14, but our data extend this observation to isotope compositions, notably the decrease of the liver δ65Cu value during aging (Fig. 1f). Isotopic aging “clocks” have been described in yeast (D/H, 13C/12C, 15N/14N,27), worm (65Cu/63Cu,28), and in human blood (65Cu/63Cu, 66Zn/64Zn29). Furthermore, metallomic or isotopic aging clocks are much easier to handle than DNA methylation clocks (performed for a subset of livers of the same animals30) which require a predetermined profile of methylation sites in the organism and organ of interest. To find metal correlations not solely driven by age, we hereafter used linear regression to correct metallomic concentrations for age effects. This correction is applied hereafter except if specified otherwise and ultra-trace elements are excluded to compare organs with the same set of metals. The concentration of major elements Mg, K, P, and S, but not Na, are positively and strongly correlated in all organs and conserved after age correction, highlighting the fundamental interplay throughout life between cationic electrolytes (Mg2+ and K+), ATP (PO4) and glutathione (S)31 (Figs. 1g and S3). Minor elements Ca and, to a lesser extent Fe, are negatively correlated to other elements in many organs (Fig. 1g).
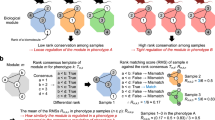
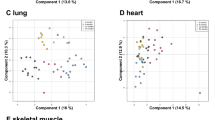
The metallome fingerprint is highly conserved across different studies
We compared our results with a recent mouse study14, which measured a smaller subset of the metallome in mouse organs at different ages. With the same set of metals and organs, the respective PCAs agree almost perfectly, demonstrating a highly conserved metallome distribution in mouse organs (Fig. 2a, b). For a given organ, age effects converged in both studies despite very different timepoints (16 timepoints at 2 months intervals14 versus 3 timepoints in this study) and different diets (NIH-3114 vs Teklad 18% protein) (Fig. 2c). After age correction, the correlation networks generated with both datasets still converge in most organs (Figs. S4 and 2d). This suggests a tight control of the metallome, reflective of the duality of metals, being bioessential at normal concentrations and toxic outside. From an evolutionary perspective, the metallome homeostasis likely evolved billions of years ago when early life had to adapt to an inorganic environment driven by ocean chemistry32. Our work reveals that not only basal metal concentrations in organs are conserved across unrelated studies, but those subtle variations are also conserved, as evidenced by the similar interaction network and reproducible response to aging. Given this, we asked whether these metal variations would relate to perturbations in other biological layers, notably the phenome, metabolome, and proteome.
a, b Robustness of PCA analysis of the mouse metallome across different data sets. a PCA for the present study. b PCA analysis for the study of Zhang et al.14 c Scatterplots of Spearman Rho correlation coefficients, and Spearman test FDR-corrected p-value calculated between metals and age in the data from Zhang et al. vs. those of the present study. The Zhang dataset measured metals at 16 timepoints at 2 months interval, as opposed to our studies’ 3 timepoints, yet trends converge towards the same age effects in the liver, muscle, and kidney. The effects of age in the heart do not match, but are very low in both studies. d Scatterplots of Spearman Rho correlation coefficients and Spearman test FDR-corrected p-value calculated between pairs of metals from Zhang et al. vs. those of the present study. To enable comparisons between the two studies, data were normalized to Na in a, b and total metal concentrations in c, d. In panels c and d, the black line and gray error band represent a linear regression and its 95% confidence interval, respectively. Source data are provided as a Source Data file.
The metallome and isotope compositions are associated with metabolic health and mitochondrial content
We recorded cardiometabolic phenotypic traits, including body weight, fat content, O2 consumption, glucose level, and blood pressure, and compared these with the metallome and δ65Cu and δ66Zn profiles through pairwise correlations (Figs. 3a, b and S5). Amongst the metallome-phenotype associations, the most significant is a negative correlation between the liver δ65Cu value and body weight (Fig. 3b), and to a lesser extent fat% and a slower return to glucose homeostasis after intraperitoneal injection (IPGTT AUC, Fig. 3b), an indicator of diabetic-like symptoms. Iron and δ65Cu are further associated with an increase in the activity of mitochondrial complex I and a reduction of the mtDNA/nuclear DNA ratio in the liver, suggesting that liver mitochondrial activity is associated to these changes in metal levels. A high liver δ65Cu is therefore associated with both younger (Fig. 1f) and metabolically healthier (Fig. 3b) animals. While increased Cu concentration33 and isotope composition4,34,35 have been previously associated with metabolic activity and growth of tumors, our study shows that δ65Cu may be a more general indicator of metabolic fitness. Importantly, these liver metal concentrations may not be causal in metabolic fitness, but rather represent a biomarker of liver health in metabolically healthy animals, as opposed to mildly obese or diabetic animals which develop liver dysfunction. Increased liver Fe further correlated with fat percentage and muscle ROS production (Fig. 3c), which are produced primarily by mitochondria during muscle exercise36. In addition, liver calcium was strongly correlated with the activity of mitochondrial complex IV. Calcium has been shown to bind and inhibit mitochondrial complex IV in vitro37, but these results suggest that interactions between Ca and complex IV may also be relevant in vivo. Kidney Fe also correlated with respiratory capacity (VO2 increase, Fig. 3c) reflecting the importance of Fe signaling in kidney erythropoietin production, a critical regulator of hemoglobin and erythrocyte production38.
a Significant correlations between metabolic phenotypes and metals in the liver and kidney. There were no significant associations in other organs except for a single one in the heart. This is consistent with the central role of liver and kidney in the control of metabolic traits. ***Adj.P < 0.001, **Adj.P < 0.01, *Adj.P < 0.05, ‘Adj.P < 0.1. b, c Dot plots of the individual correlations of δ65Cu (b) and Fe (c) with indicators of obesity (body weight, fat %), diabetes (Intra-peritoneal glucose tolerance test, area under the curve, IPGTT AUC), and muscle activity (Muscle ROS) and running performance (VO2 increase), as well as the activity of mitochondrial complex I. The subscripts (ac) stand for “age corrected”. In all panels, Rho indicates the Spearman Rho coefficient, and Adj.P is computed by a moderated t-test (limma), two-sided and FDR-corrected with age as a covariate. In panels b and c, the black line and gray error band represent a simple linear regression and its 95% confidence interval, respectively. Source data are provided as a Source Data file.
The metallome and isotope compositions fingerprints integrate with overlapping biological pathways in the proteome and metabolome
To examine the interplay between metallomics and other omics layers, we used previously published metabolomics39 and new proteomics analyses performed on liver samples from the same animals. A Metabolite Set Enrichment Analysis (MSEA) and Gene Set Enrichment Analysis (GSEA) based on correlations between the metallome, the metabolome (Fig. 4a), and the proteome (Fig. 4b) define four groups of correlated metals. Group I (S, Mg, P, Rb, K, Cu, Zn, and Mn) displays strong positive normalized enrichment scores (NES) for amino, organic, and carboxylic acids and derivatives and negative NES for fatty acids, lipids, and derivatives, with opposite enrichments in Group II (Na and Ca) (Fig. 4a). This pattern is independently confirmed by the GSEA on hallmark gene sets in the proteome, where groups I and II oppositely correlate with adipogenesis and fatty acid metabolism (Fig. 4b). The MSEA for cellular locations indicates that group I associates with cytoplasmic components but negatively correlates with membrane components (Fig. 4a). Group I also negatively correlates with the mitochondria (Fig. S8), in accordance with the Hallmark GSEA concerning the oxidative phosphorylation pathway (OXPHOS, Fig. 4b). Group III consists of δ65Cu only and is positively associated for amino, organic and carboxylic acids and derivatives in the chemical classes MSEA (Fig. 4a). The previously observed negative phenotypic correlation between the δ65Cu value and fat % (Fig. 3b) is strengthened by the Hallmark GSEA (Fig. 4b) implicating adipogenesis. Group IV (Fe and δ66Zn) had comparably fewer enrichments with the MSEA (Fig. 4a) but reveals many significant enrichments with the GSEA (Figs. 4b and S8). This group associates with mitochondrial content, in accordance with the role of Fe in heme synthesis, electron transport and oxidative phosphorylation, and the mitochondrial biosynthesis of the critical Fe-S cluster proteins40,41. The discovery that the δ66Zn value varies in opposition to Zn concentration, notably with mitochondrial proteins (GO CC, Figs. 4b and S8), emphasizes the added value of determining stable isotope composition in addition to concentration to highlight metal-based molecular pathways. The KEGG GSEA (Fig. S8) links δ66Zn and neurodegenerative diseases, predicting a connection that was experimentally confirmed in the APPswe/ PSEN1dE9 mouse model of Alzheimer disease42,43.
a, b Metabolites and proteins were sorted on their correlation with individual metals in the liver, then metabolite or gene -set enrichment analyses (MSEA, (a), GSEA, (b)) were performed. For each metal, the top three gene or metabolite sets are pictured. FDR-corrected p-values.**Adj.P < 0.01, *Adj.P < 0.05, ‘Adj.P < 0.1 (c) Network representation, combining the most significant correlations between metals, between metals and phenotypes (Spearman Rho and limma test, both), as well as the metabolite and gene set enrichment analyses. All tests were two-sided when applicable and p-values are FDR corrected. To limit the number of nodes, only nodes with very significant associations are pictured (Adj.P < 0.01), but all edges with Adj.P < 0.05 are drawn. Source data are provided as a Source Data file.
We summarize the results of the study in a network analysis gathering correlations between metals, phenotypes, as well as the MSEA and GSEA results in the liver (Fig. 4c). The resulting network recapitulates all the above observations (e.g. δ65Cu and body weight, Fe and OXPHOS) and shows that the metallome significantly interacts with all the studied omic layers. The network highlights the similar roles of K and Rb which strengthen the postulated status of Rb as a proxy for K metabolism.
Perspectives
Overall, our findings establish that the metallome and the isotopic compositions are biologically relevant and highly integrated omic layers with an unprecedented level of conservation in both basal signature and evolution with age. Subtle variations in metal concentrations are conserved across studies and associated with markers of metabolic health at both the physiological (body weight, fat percentage, and insulin resistance in the IPGTT) and biochemical levels (OXPHOS, lipid synthesis among others). A limitation of this study is that it is mostly based on correlations, and we do not imply that alterations in metals are causal in the phenotypes observed. However, the high reproducibility of metallomics suggests that changes in metal concentration may represent a source of reliable and affordable biomarkers of organismal, tissular, cellular, and subcellular aging and metabolism.
Methods
Mouse sample collection
All animal experiments were performed according to Swiss ethical guidelines and approved by the local animal experimentation committee of the Canton de Vaud under license 2172. Male C57BL/6J mice (Janvier, St Berthevin, France) were fed with a standard chow diet (containing 18% protein, 50% carbohydrate, and 6.0% fat; Harlan Laboratories, Madison, WI, USA) at the Laboratory of Integrative Systems Physiology (LIPS) at EPFL (Ecole Polytechnique Fédérale de Lausanne). The mice were kept with a 12-h light cycle and temperature was regulated at 23 °C and 40–60% humidity. After in vivo phenotyping, organs (i.e. liver, brain, heart, muscle (i.e. quadriceps), and kidney) were collected at three different time-points in their life i.e. 6 months (mature adult), 16 months (middle age) and 24 months (old). The organs were flash frozen in liquid nitrogen (N2), and stored at −80 °C, to promote their conservation before chemical and metabolomics analyses.
In vivo phenotyping
All phenotype data used in this paper was previously published in Houtkooper et al.39, but a brief description of the procedures is provided here. Body composition was determined by Echo-MRI (Echo Medical Systems, Houston, TX, USA) and oxygen consumption (VO2), respiratory exchange ratios (RER), activity levels and activity were monitored by indirect calorimetry using the comprehensive laboratory animal monitoring system (CLAMS) (Columbus Instruments, Columbus, OH, USA). Glucose tolerance was analyzed by measuring blood glucose and insulin following intraperitoneal injection of 2 g/kg glucose after an overnight fast. Maximum respiration potential (VO2max) was determined in a metabolic treadmill (Columbus instruments, Columbus, OH, USA) with an incremental speed protocol. During the run, VO2 and VCO2 are measured. The experiment is stopped when mice are exhausted, RER is above 1 for more than one interval, or the VO2 reaches a clear plateau. Liver mitochondrial complex I and IV activity (Mitosciences, Eugene, OR, USA), citrate synthase (Sigma) were determined according to the manufacturer’s protocols. Liver and muscle ROS were determined by 4-HNE detection in PBS tissue lysates using an ELISA-based method (Cell Biolabs, San Diego, CA, USA).
Analyses of major and trace element concentrations
All the chemical analyses were carried out in a clean laboratory below laminar flow clean hoods using ultra-pure water at 18.2 MΩ.cm and acids that were doubly distilled to ensure low trace element contents and avoid any exogenous contaminations. Blanks were also run to quantify and integrate potential contamination from the material and/or acids used to perform experiments. The samples were crushed under liquid N2, freeze-dried, weighted, and then dissolved in a concentrated HNO3-H2O2 30% mixture in Savillex beakers for about 72 h at 120 °C. Once dissolved, major (Ca, K, Na, P, S, and Mg) and trace elements (Mn, Co, Cu, Zn, Fe, Se, Rb, Mo, and Cd) were measured using the ICP-AES, iCAP 6000 Radial and the quadrupole ICP-MS Thermo iCap-Q respectively at the Ecole Normale Supérieure (ENS) of Lyon. The concentrations were calculated using calibration curves determined based on multi-elemental solutions (SP-33MS for trace element and in-house solution made from mono-elemental solutions for major element) following the procedure detailed in Garçon et al.44. Both accuracy and reproducibility were monitored by complete duplicate analyses, replication of an in-house (sheep plasma; OEP) and certified standards (bovine liver, SRM-1577c) as well as re-run analyses over the course of an analytical session. Given our long-term reproducibility on the in-house/certified standards and duplicate analyses, the 2 standard deviations (2 SD) analytical uncertainty adopted in this study for both trace and major concentrations are on average better than 10% (Tables S1 and S2).
Analyses of copper and zinc isotopic compositions
Copper and zinc isotopic compositions were measured at the Ecole Normale Supérieure (ENS) of Lyon following the procedure described by Maréchal et al.45. Briefly, before each isotopic measurement, the sample solutions were purified by ion-exchange chromatography using quartz columns filled with 1.8 mL of Bio-Rad AGMP-1 (100–200 mesh) anion-exchange resin. Both copper and zinc were successively eluted with 20 mL of HCl 7 N + 0.001% H2O2 and 10 mL of HNO3 0.5 M, respectively. On the day of the analyses, Zn and Cu purified solutions, previously evaporated to dryness, were dissolved in a Cu (Cu SRM 976, National Institute of Standards and Technology, Gaithersburg, MD, USA) or Zn-doped solution (Zn JMC 3-0749 L, Johnson Matthey Royston, UK) respectively to reach a Zn or Cu sample concentration of about 300 ppb, which is similar to the concentration of the standard solution that was run between each sample.
Copper and Zn isotopic compositions are expressed as:
and
Copper and Zn isotopic compositions were measured on a Nu Plasma (Nu 500) MC-ICP-MS in wet plasma conditions. Zn JMC 3-0749L (also called JMC-Lyon; Johnson Matthey, Royston, UK) and Cu SRM 976 were used as reference standards for δ66Zn and δ65Cu, respectively. Instrumental mass fractionation was corrected with an exponential law using an elemental-doping method and instrumental drift over time was controlled with standard sample bracketing45
The accuracy of isotopic compositions was assessed by the analysis of in-house and commercial standard solutions (i.e. sheep plasma; OEP and bovine liver SRM-1577c certified reference material46) during each analytical sequence. For Zn, the average δ66Zn value measured is +0.76 ± 0.08‰ (n = 3, 2 SD) for OEP and −0.18 ± 0.05‰ (n = 4, 2 SD) for SRM-1577c which is in good agreement with the average values 0.73 ± 0.09‰ (n = 17, 2 SD) for OEP and −0.19 ± 0.06‰ (n = 20, 2 SD) for SRM-1577c (4). For Cu, we measured the OEP δ65Cu value at −1.10 ± 0.15‰ (n = 3, 2 SD) and the SRM-1577c δ65Cu value at +0.43 ± 0.04‰ (n = 4, 2 SD) which is also in good agreement with previously estimated values (δ65CuOEP = −1.14 ± 0.13 ‰ (2 s, n = 35) and δ65Cu1577c = +0.37 ± 0.14 ‰ (n = 19, 2 SD, (4), Table S1). Based on re-run samples, complete duplicate analyses and standard measurements, we estimate the precision of our measurements at ±0.12‰ (2 SD). The long-term precision based on the repeated measurements of standard Zn JMC 3-0749L and Cu SRM 976 alone is however better than ±0.06‰ (n = 140, 2 SD).
Metabolomics
The liver metabolomics was performed by Metabolon (Durham, NC, USA) and has already been published47. Briefly, sample preparation was conducted using a proprietary series of organic and aqueous extractions to remove the protein fraction while allowing maximum recovery of small molecules. The extracted samples were split into equal parts for analysis on the GC/MS and LC-MS/MS platforms. For LC-MS/MS, samples were split in two aliquots that were either analyzed in positive (acidic solvent) or negative (basic solvent) ionization mode. GC-MS was performed on bistrimethyl-silyl-triflouroacetamide derivatized samples in a 5% phenyl GC column.
Proteomics
Proteomics was performed as described in Yu et al.48. Briefly, total proteome was isolated using a RIPA-M buffer (pH 7.5; nonidet NP-40, 0.1% sodium doexycholate, 150 mM NaCl, 1 mM EDTA, 50 mM Tris, protease inhibitor cocktail, 10 mM NaF, 5 mM 2-glycerophosphate) and lysed with a 8 M urea buffer (pH 8.1, 75 mM NaCl, 10 mM NaF, 5 mM 2-glycerophosphate; 8 M urea; protease inhibitor cocktail). Tissues were ground using a pestle grinder in the Ripa-M buffer before the samples were lysed in the urea buffer. After proteins were isolated, samples were trypsin digested overnight in the dark (~22 h @ 37 °C in a mild shaker) in a urea buffer with dithioethreitol and indole-3-acetic acid. After trypsinization, samples were passed through a C18 column and eluted in a 2% ACN solution with 0.1% FA. Samples were then run on a TripleTOF 5600 in SWATH mode. The search library used was generated from that of an earlier study49. For the data processing, we removed decoy peptides and peptides mapping only to reverse proteins, then summed raw intensities over different charge states and chemical modifications. The resulting peptide-level intensities were log2 transformed and normalized by robust linear regression normalization, separately for each organ. We then removed peptides that are identified in only one sample, as well as proteins identified by a single unique peptide sequence. Finally, peptide-level intensities were summarized to the protein level with the MsqRobSum R package50, (proteins were grouped by gene annotations).
Data analysis
Outlier measurements were removed using the ‘normal’ method (differences between each value and the mean divided by standard deviation) through the R package ‘outliers’. Less than 5% of measurements were removed this way. All correlation analyses were performed by parallel linear regression using the ‘limma’ R package using age as a covariate51. P-values were corrected for multiple testing through the Benjamini–Hochberg false discovery rate method. Pearson correlation coefficients were computed separately through the ‘cor.test’ function. Age-corrected values were computed by estimating the effect of age through a linear model value ~age and extracting the intercept + residuals of the model. Those values were used for visualization only, but statistical analyses were performed using age as a covariate instead.
MSEA
In order to use the most complete and up-to-date repository of metabolite information, we based our analysis on the Human Metabolome Database (HMDB). We downloaded the database and parsed information about biological processes and cellular components from the relevant entries in the database, then generated a search file for the ClusterProfiler R package52. For MSEA, metabolites were sorted based on their degree of correlation with each metal (calculated with limma package, see ‘analysis’ section) using the following formula: −log10(P-value)* Pearson correlation coefficient. Metabolite Set enrichment analysis (MSEA) was performed using the ClusterProfiler package in R software.
GSEA
Gene set databases were downloaded from the Molecular Signatures Database (MSigDB) v7.2 through the MSigDRr R package. Similar to GSEA, all proteins were sorted by −log10(P-value)* Pearson correlation coefficient and GSEA was performed with the ClusterProfiler R package.
Networks
All network representations show the correlation analyzes and MSEA/GSEA results outlined above. Only correlations with Adj. P-values < 0.005 were drawn. Network visualizations were generated using the igraph and ggraph R packages.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Raw metallomic data are included in this study as supplementary data 1. Proteomics has been submitted to the PRIDE repository under the identifier PXD01114253. Metabolomics data have been previously published and are available in Houtkooper et al.39. In addition, source data files are provided for every figure panel.
Code availability
Data analysis was performed in R software version 3.5.2. The following open-source packages were used in analysis and figure generation: MsqRobSum: proteomics; limma: differential analyzes and parallel linear regressions; ClusterProfiler: Gene and Metabolite set enrichment analyzes; ggplot2: figure generation; igraph, ggraph: network visualizations. All code used in the paper is available on request.
References
Lobinski, R., Becker, J. S., Haraguchi, H. & Sarkar, B. Metallomics: Guidelines for terminology and critical evaluation of analytical chemistry approaches (IUPAC Technical Report). Pure Appl. Chem. 82, 493–504 (2010).
Bigeleisen, J. & Mayer, M. G. Calculation of equilibrium constants for isotopic exchange reactions. J. Chem. Phys. 15, 261–267 (1947).
Bigeleisen, J. The relative reaction velocities of isotopic molecules. J. Chem. Phys. 17, 675–678 (1949).
Balter, V. et al. Natural variations of copper and sulfur stable isotopes in blood of hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA 112, 982–985 (2015).
Walczyk, T. & von Blanckenburg, F. Natural iron isotope variations in human blood. Science 295, 2065–2066 (2002).
Morgan, J. L. L. et al. Rapidly assessing changes in bone mineral balance using natural stable calcium isotopes. Proc. Natl. Acad. Sci. USA 109, 9989–9994 (2012).
Lahner, B. et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21, 1215–1221 (2003).
Salt, D. E., Baxter, I. & Lahner, B. Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 59, 709–733 (2008).
Stich, B. et al. The maize shoot ionome: Its interaction partners, predictive power, and genetic determinants. Plant Cell Environ. 43, 2095–2111 (2020).
Eide, D. J. et al. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 6, R77 (2005).
Malinouski, M. et al. Genome-wide RNAi ionomics screen reveals new genes and regulation of human trace element metabolism. Nat. Commun. 5, 3301 (2014).
Balter, V. et al. Contrasting Cu, Fe, and Zn isotopic patterns in organs and body fluids of mice and sheep, with emphasis on cellular fractionation. Metallomics 5, 1470–1482 (2013).
Ma, S. et al. Organization of the mammalian ionome according to organ origin, lineage specialization, and longevity. Cell Rep. 13, 1319–1326 (2015).
Zhang, B., Podolskiy, D. I., Mariotti, M., Seravalli, J. & Gladyshev, V. N. Systematic age-, organ-, and diet-associated ionome remodeling and the development of ionomic aging clocks. Aging Cell 19, e13119 (2020).
Balter, V. et al. Bodily variability of zinc natural isotope abundances in sheep. Rapid Commun. Mass Spectrom. 24, 605–612 (2010).
Jaouen, K. et al. Dynamic homeostasis modeling of Zn isotope ratios in the human body. Metallomics 11, 1049–1059 (2019).
Albarède, F., Télouk, P. & Balter, V. Medical applications of isotope metallomics. Rev. Mineral. Geochem. 82, 851–885 (2017).
Crabb, E. & Moore, E. Metals and Life (Royal Society of Chemistry, 2010).
Hahn, P. et al. Age-dependent and gender-specific changes in mouse tissue iron by strain. Exp. Gerontol. 44, 594–600 (2009).
Massie, H. R., Aiello, V. R. & Banziger, V. Iron accumulation and lipid peroxidation in aging C57BL/6J mice. Exp. Gerontol. 18, 277–285 (1983).
Morita, A., Kimura, M. & Itokawa, Y. The effect of aging on the mineral status of female mice. Biol. Trace Elem. Res. 42, 165–177 (1994).
Takahashi, S. et al. Age-related changes in the concentrations of major and trace elements in the brain of rats and mice. Biol. Trace Elem. Res. 80, 145–158 (2001).
Barnham, K. J. & Bush, A. I. Biological metals and metal-targeting compounds in major neurodegenerative diseases. Chem. Soc. Rev. 43, 6727–6749 (2014).
Barnham, K. J., Masters, C. L. & Bush, A. I. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 3, 205–214 (2004).
Godt, J. et al. The toxicity of cadmium and resulting hazards for human health. J. Occup. Med. Toxicol. Lond. Engl. 1, 22 (2006).
Fieve, R. R. et al. Rubidium: biochemical, behavioral, and metabolic studies in humans. Am. J. Psychiatry 130, 55–61 (1973).
Li, X. & Snyder, M. P. Yeast longevity promoted by reversing aging-associated decline in heavy isotope content. Npj Aging Mech. Dis. 2, 1–10 (2016).
Sauzéat, L., Laurençon, A. & Balter, V. Metallome evolution in ageing C. elegans and a copper stable isotope perspective. Metallomics 10, 496–503 (2018).
Jaouen, K. et al. Is aging recorded in blood Cu and Zn isotope compositions? Metallomics 5, 1016–1024 (2013).
Bou Sleiman, M. et al. The gene-regulatory footprint of aging highlights conserved central regulators. Cell Rep. 32, 108203 (2020).
Bennett, B. D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009).
Anbar, A. D. & Knoll, A. H. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297, 1137–1142 (2002).
Ishida, S., Andreux, P., Poitry-Yamate, C., Auwerx, J. & Hanahan, D. Bioavailable copper modulates oxidative phosphorylation and growth of tumors. Proc. Natl. Acad. Sci. USA 110, 19507–19512 (2013).
Lobo, L. et al. Elemental and isotopic analysis of oral squamous cell carcinoma tissues using sector-field and multi-collector ICP-mass spectrometry. Talanta 165, 92–97 (2017).
Télouk, P. et al. Copper isotope effect in serum of cancer patients. A pilot study. Met. Integr. Biometal Sci. 7, 299–308 (2015).
Barbieri, E. & Sestili, P. Reactive oxygen species in skeletal muscle signaling. J. Signal Transduct. 2012, 982794 (2012).
Vygodina, T., Kirichenko, A. & Konstantinov, A. A. Direct regulation of cytochrome c oxidase by calcium ions. PLoS ONE 8, e74436 (2013).
Adamson, J. Erythropoietin, iron metabolism, and red blood cell production. Semin. Hematol. 33, 5–7 (1996).
Houtkooper, R. H. et al. The metabolic footprint of aging in mice. Sci. Rep. 1, 134 (2011).
Richardson, D. R. et al. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. USA 107, 10775–10782 (2010).
Braymer, J. J. & Lill, R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 292, 12754–12763 (2017).
Moynier, F., Foriel, J., Shaw, A.S. & Le Borgne, M. Distribution of Zn isotopes during Alzheimer’s disease. Geochem. Perspect. Lett. https://www.geochemicalperspectivesletters.org/article1717/ (2017).
Büchl, A., Hawkesworth, C. J., Ragnarsdottir, K. V. & Brown, D. R. Re-partitioning of Cu and Zn isotopes by modified protein expression. Geochem. Trans. 9, 11 (2008).
Garçon, M. et al. Nitrile, latex, neoprene and vinyl gloves: a primary source of contamination for trace element and Zn isotopic analyses in geological and biological samples. Geostand. Geoanal. Res. 41, 367–380 (2017).
Maréchal, C. N., Télouk, P. & Albarède, F. Precise analysis of copper and zinc isotopic compositions by plasma-source mass spectrometry. Chem. Geol. https://www.sciencedirect.com/science/article/pii/S0009254198001910 (1999).
Sauzéat, L. et al. Inter-comparison of stable iron, copper and zinc isotopic compositions in six reference materials of biological origin. Talanta https://www.sciencedirect.com/science/article/pii/S0039914020308675?dgcid=rss_sd_all (2021).
Houtkooper, R. H. et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457 (2013).
Wu, Y., Williams, E. G. & Aebersold, R. Application of SWATH proteomics to mouse biology. Curr. Protoc. Mouse Biol. 7, 130–143 (2017).
Williams, E. G. et al. Quantifying and localizing the mitochondrial proteome across five tissues in a mouse population. Mol. Cell. Proteomics 17, 1766–1777 (2018).
Sticker, A., Goeminne, L., Martens, L. & Clement, L. Robust summarization and inference in proteome-wide label-free quantification. Mol. Cell. Proteomics 19, 1209–1219 (2020).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMIICS 16, 284–287 (2012).
Williams, E., Auwerx, J. & Aebersold, R. Multi-omics analyses revealed conserved aging signatures in mice. Project PXD011142. https://www.ebi.ac.uk/pride/archive/projects/PXD011142.
Acknowledgements
We thank all members of J. Auwerx’s laboratory for helpful discussions. This project has received financial support from the CNRS through the MITI interdisciplinary programs, the Bullukian Foundation, the École Polytechnique Fédérale de Lausanne (EPFL), European Research Council (ERC-AdG-787702) and Swiss National Science Foundation (31003A_179435) and a Global Research Laboratory grant from the National Research Foundation of Korea (2017K1A1A2013124). L.S. was supported by EPFL and ENS de Lyon convention thanks to P. Gillet. L.J.E.G. was supported by the European Union Horizon 2020 Marie Skłodowska-Curie Individual Fellowship “AmyloAge” (896042).
Author information
Authors and Affiliations
Contributions
V.B. and J.A. conceived the project. R.H performed mouse clinical phenotyping and tissue collection. L.S. performed the metallomic measurements. R.A. and E.W. generated the proteomics dataset. J.D.M., V.B., and L.J.E.G analyzed the data with the help of L.S. and P.J.; V.B and J.D.M. wrote the manuscript with comments from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alessandro Ori and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Morel, JD., Sauzéat, L., Goeminne, L.J.E. et al. The mouse metallomic landscape of aging and metabolism. Nat Commun 13, 607 (2022). https://doi.org/10.1038/s41467-022-28060-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-28060-x
This article is cited by
-
Metal-binding amino acid ligands commonly found in metalloproteins differentially fractionate copper isotopes
Scientific Reports (2024)
-
Zinc and iron dynamics in human islet amyloid polypeptide-induced diabetes mouse model
Scientific Reports (2023)
-
Translational toxicology of metal(loid) species: linking their bioinorganic chemistry in the bloodstream to organ damage onset
BioMetals (2023)
-
The ionome and proteome landscape of aging in laying hens and relation to egg white quality
Science China Life Sciences (2023)
-
Magnesium stable isotope composition, but not concentration, responds to obesity and early insulin-resistant conditions in minipig
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.