Abstract
KdpFABC, a high-affinity K+ pump, combines the ion channel KdpA and the P-type ATPase KdpB to secure survival at K+ limitation. Here, we apply a combination of cryo-EM, biochemical assays, and MD simulations to illuminate the mechanisms underlying transport and the coupling to ATP hydrolysis. We show that ions are transported via an intersubunit tunnel through KdpA and KdpB. At the subunit interface, the tunnel is constricted by a phenylalanine, which, by polarized cation-π stacking, controls K+ entry into the canonical substrate binding site (CBS) of KdpB. Within the CBS, ATPase coupling is mediated by the charge distribution between an aspartate and a lysine. Interestingly, individual elements of the ion translocation mechanism of KdpFABC identified here are conserved among a wide variety of P-type ATPases from different families. This leads us to the hypothesis that KdpB might represent an early descendant of a common ancestor of cation pumps.
Similar content being viewed by others
Introduction
KdpFABC (K+-dependent P-type ATPase) is a K+ pump that is responsible for high-affinity electrogenic K+ uptake into prokaryotes at very low extracellular K+ concentrations1,2,3. The heterotetrameric complex combines a P-type ATPase (KdpB) with a channel-like subunit (KdpA) from the superfamily of K+ transporters (SKT)4,5. These subunits are augmented by KdpF, a single transmembrane (TM) helix which stabilizes the complex6, and KdpC, which has been suggested to influence substrate affinity7, although its exact function remains elusive. Its chimeric architecture makes KdpFABC unique among K+ transporters, and ensures the active transport of K+ with an apparent affinity of 2 µM and with high substrate specificity2,8,9,10. To fulfill this physiological role, both KdpA and KdpB seem to diverge substantially from other members of their families.
The channel-like subunit KdpA consists of four nonidentical, covalently linked membrane-pore-membrane (MPM) motifs, referred to as domains 1–4 (D1–D4), which are arranged in a pseudo-four-fold symmetrical pore. An additional transmembrane helix each is fused at the N and C terminus. KdpA, like other members of the SKT, contains a selectivity filter (SF) that is less conserved than the canonical TVGYG motif of K+ channels. This has been proposed to reduce the number of K+ binding sites and suggests a lower ion selectivity11,12. However, unlike the SKT members TrkH and KtrB, ion transport by KdpFABC was shown to be highly selective for K+ over Na+, and ATPase activation by ammonium or Rb+ was only possible upon mutation of the KdpA SF region9,10. Furthermore, the gating region of the ion channels KtrB and TrkH is not conserved in KdpA. In the latter, the intramembrane loop in D3M2 is much less polar and the highly conserved arginine is not present in D4M213.
KdpB is a 7-TM P-type ATPase. Like all other P-type ATPases, KdpB alternates between outward- and inward-open conformations in the so-called Post-Albers cycle, which features states E1 and E2, and the phosphorylated intermediates E1-P and E2-P14,15. These states are characterized by large rearrangements of the cytosolic domains (N—nucleotide binding, P—phosphorylation, and A—actuator) of KdpB, which are responsible for nucleotide binding and hydrolysis12,16,17. Dephosphorylation is mediated by a TGES motif in the A domain, which is widely conserved among cation P-type ATPases18,19,20. A unique feature of the TGES motif of KdpB is that it is subject to an inhibitory serine phosphorylation (Escherichia coli (Ec)KdpBS162). This phosphorylation has been shown to prevent excessive K+ uptake when external K+ is high, indicating that it limits uptake by KdpFABC to conditions where it is physiologically necessary16,21. Furthermore, KdpB is the smallest known P-type ATPase22, and was long assumed to be unable to transport substrates itself. Instead, it was suggested to solely provide the energy for ion translocation through the channel-like subunit KdpA23. However, the spatial separation of the subunits purported to be responsible for ATP hydrolysis and ion transport posed a conundrum as to how the two processes are coupled.
Several recent structural studies of EcKdpFABC have provided possible explanations of how the unique structural features of both KdpA and KdpB effect K+ transport. The first X-ray structure of KdpFABC solved in an E1 state led to the identification of an aqueous tunnel within the complex, running along the membrane plane and extending from the SF in KdpA to the P-type ATPase canonical substrate binding site (CBS) in KdpB16. Initially, the coupling helix model was proposed, in which the presence of K+ ions in the SF of KdpA is communicated to KdpB via a Grotthuss (also known as a proton wire) mechanism running through the intersubunit tunnel16. KdpB hydrolyzes ATP, undergoes a conformational change, and pulls open the intramembrane gate in the channel subunit via a coupling helix, thereby enabling K+ flux through KdpA. The periplasmically oriented KdpC could function as external gate.
For this model to work, a salt bridge between KdpB and KdpA is required to pull on the coupling helix. This salt bridge has, however, been shown to be dispensable, as it was broken in a subsequent cryo-EM structure of KdpFABC in an E2 state17. In this structure, virtually no conformational changes in KdpA and KdpC were observed when compared to the E1 state. Instead, the intersubunit tunnel was restricted at the KdpA–KdpB interface, while a new half-channel in KdpB from the CBS to the cytosol opened. This is accomplished by a significant reorientation of the positively charged KdpBK586 in the CBS and the surrounding TM helices. Finally, a cryo-EM map of KdpFABC in an E1 state contained several nonprotein densities within the intersubunit tunnel, which were suggested to be K+ ions.
Based on these structural insights, the intersubunit tunnel translocation model was suggested17: in the E1 state, extracellular K+ ions are forwarded from the SF in KdpA through the intersubunit tunnel to the CBS in KdpB, where they trigger ATP hydrolysis and phosphorylation of the catalytic KdpBD307. The ensuing E1-P/E2-P transition is associated with a reorientation of KdpBK586, which pushes K+ out of the CBS and simultaneously acts as a built-in counterion to stabilize the E2/E1 transition. Recent cryo-EM structures extend the model by suggesting a displacement of the ion from the CBS into an adjacent low-affinity site in the E2-P state, from where ion release into the cytoplasm is favored12.
Here, we decipher the molecular basis of the transport pathway of KdpFABC by addressing how K+ ions are forwarded through the tunnel, how they bind to the CBS, and the mechanisms by which ATP hydrolysis and the subsequent E1-P/E2-P transition are triggered. We present two cryo-EM structures of KdpFABC in an E1·ATP state, loaded with K+ and Rb+ ions, respectively, allowing a more unambiguous assignment of ion positions in the complex. Further, we characterize the dynamics of ion coordination and translocation through the intersubunit tunnel by molecular dynamics (MD) simulations. Finally, we combine biochemical and MD approaches to identify three conserved residues responsible for ATPase coupling, ion propagation through the intersubunit tunnel into the CBS, and regulation of the rate-limiting step of turnover, providing the foundation for K+ transport by KdpFABC.
Results
Structures of K+- and Rb+-loaded KdpFABC in an E1·ATP state
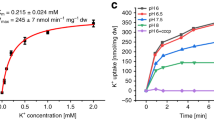
In various cryo-EM structures of KdpFABC, nonprotein densities have been observed along the intersubunit tunnel. Although the densities could not be unambiguously assigned, they were suggested to represent K+ ions or water molecules12,16,17. Due to its higher atomic number, Rb+ (Z = 37) scatters electrons more strongly than K+ (Z = 19) by an expected factor of 2.524,25,26. We set out to exploit this as a strategy to assign substrate ions with more confidence. To this end, we determined cryo-EM structures of the complex loaded with K+ and Rb+, both arrested in an E1·ATP state. In the K+-loaded sample, we used the mutation KdpBD307N, which precludes ATP hydrolysis, to stall the complex in the E1 state. To incorporate Rb+, we employed variant KdpFAG232DBS162AC, in which a central glycine (KdpAG232) of the KdpA SF is mutated to allow passage of Rb+9. K+ ions co-purified in the intersubunit tunnel of the variant were exchanged for Rb+ by applying turnover conditions. Here, the KdpBS162A mutation prevented the inhibitory phosphorylation in the A domain, enabling turnover.
In both samples, the complex was trapped by the addition of the nonhydrolysable ATP analog AMPPCP prior to cryo-EM sample preparation. The structures were determined at a global resolution of 3.1 and 3.2 Å in the K+- and Rb+-loaded samples, respectively, with higher local resolution in the transmembrane region. Both structures are nearly identical and adopt an E1·ATP conformation (Fig. 1a, Supplementary Figs. 1–6, Supplementary Table 1). The nucleotide AMPPCP is coordinated between the N and P domains, reflecting the common catalytic ATP binding mode of P-type ATPases27 (Supplementary Fig. 6b, c). This state shows an outward-open half-channel that reaches from the KdpA SF to the CBS in KdpB (Fig. 1a). The tunnel is sufficiently wide to accommodate either K+ or Rb+ ions along its entire length. There is a constriction point at the SF in KdpA, where K+ is dehydrated, and a second constriction immediately before the CBS (Fig. 1b, Supplementary Fig. 6d). This constriction is mainly formed by a single residue, namely KdpBF232 (Fig. 1c).
a Structural model of E1·ATP KdpFABD307NC in ribbon representation. The following color code is retained for all following figures unless otherwise specified: KdpA: green, KdpC: purple, KdpF: cyan, KdpB: TMD sand, N domain: red, P domain: blue, and A domain: yellow. Here, the KdpA pore entrance at the SF and intersubunit tunnel leading to the CBS are visualized as a pink surface. b Radius of the intersubunit tunnel along its length, from below the KdpA SF to the KdpB CBS. The tunnel is wide enough to accommodate K+ (r = 1.4 Å, dashed line) along nearly its entire length, with a significant constriction immediately before the CBS. c Top view of the constriction in the intersubunit tunnel, which is mediated mainly by KdpBF232 as it protrudes into the tunnel.
Cardiolipin aids the E1-P/E2-P transition in KdpFABC
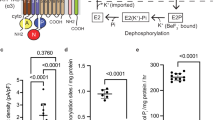
In both structures, we identify two cardiolipin (CL) molecules bound to KdpFABC (Fig. 2a, Supplementary Fig. 6e, Supplementary Fig. 7a, b). The assignment as cardiolipin in these positions is strongly supported by coarse-grained MD simulations showing preferred CL binding (Fig. 2b, Supplementary Fig. 7c). CL binding at the KdpA–KdpB interface (CL1) is principally coordinated by two basic residues (KdpAR278 and KdpBR651), a glycine residue, a polar residue and an aromatic residue (KdpAG524, KdpAH523, KdpAW285) making it a typical high-affinity E. coli CL binding site28. CL1 appears to be of integral structural importance, as evidenced by the loss of KdpFABC in vivo when the binding site is mutated (Fig. 2c, Supplementary Fig. 7e). One hydrophobic chain of CL1 reaches towards the intersubunit tunnel, lying immediately opposite KdpBF232 (Supplementary Fig. 7d). Since CL specifically stimulates the ATPase activity of KdpFABC, it may support the transition to the E2 conformation, as previously proposed12 (Fig. 2d).
a Two identified cardiolipin (CL) molecules (gray) bound to KdpFABC. A hydrocarbon tail of CL1 (dark gray) extends into the complex at the KdpA/KdpB interface. b Pooled CL contact probabilities of selected residues, from five representative structures in various E1 and E2 conformations (5MRW, 6HRA, 6HRB, 7BGY, and 7NNL), measured as proportion of frames in a coarse-grained MD simulation the CL and the respective residue are in contact, i.e., within a 0.6 nm cutoff distance. The concentration of CL in the membrane (10%) is shown as a dotted line, indicating a very high accumulation of CL interactions at these residues. Data points represent the average and error bars the standard deviation from 25 independent simulations (5 each per structure). c Anti-His Western Blot of whole-cell samples of E. coli LB2003 cells carrying KdpFABC variants, detecting His-tagged KdpC. The simultaneous knockout of residues coordinating CL1 in KdpFAR278ABR651aC abolishes protein production, showing the structural importance of CL binding for complex stability. This result was reproducible in a biological duplicate. d CL-dependent ATPase activity of KdpFABC. Rising CL conc. (0–250 µM) stimulate ATPase activity up to two fold, while additions of PE, PG, and PC do not change the ATPase activity of KdpFABC. Increased ATPase activity indicates an acceleration of the rate-limiting E1/E2 transition. Data points represent the average and error bars the standard deviation from a technical triplicate.
Ions are transported through the intersubunit tunnel
Both cryo-EM structures feature multiple distinct nonprotein densities filling the entire length of the outward-open half-channel, from the SF in KdpA to the CBS in KdpB (Fig. 3a). The individual positions in both structures closely overlap (Fig. 3a, Supplementary Fig. 9a). Compared to our previous E1 cryo-EM structure 6HRA, a higher number of densities is observed within the K+-loaded tunnel, which we attribute to the increased K+ concentration used for sample preparation (50 mM vs. 1 mM) and the improved resolution of the transmembrane domain17 (Supplementary Fig. 8a). Potential coordinating residues were identified for the majority of these positions (Supplementary Figs. 8b, 9b, Supplementary Table 2), and the tunnel is wide enough to accommodate additional waters for solvation. Together, these factors already support the assignment of the additional densities as ions.
a Potential ions in K+- and Rb+-loaded E1·ATP KdpFABC within the intersubunit tunnel, from the KdpA SF (right) to the KdpB CBS (left). Based on the high ion concentrations in each sample, densities were assigned as K+ and Rb+ ions, respectively. K+ visualized in purple, Rb+ in turquoise. b, c Comparison of densities in the CBS and in SF position S3, respectively, between the K+- and Rb+-loaded maps when low-pass filtered to 3.2 Å and displayed at the same contour level (σ). Densities in the Rb+-loaded structure (turquoise) are stronger than in the K+-loaded structure (purple), suggesting that they correspond to ions and that Rb+ is integrated into the complex. Contour levels are indicated in the top right corner of each panel. d Quantification of the effect of Rb+ substitution in the CBS and SF S3. Densities were low-pass filtered to 3.2 Å, normalized to the K+-loaded structure for each position, and indicate a ca. two-fold signal increase in the Rb+-loaded structure in both positions. For the full, non-normalized comparison of all ion densities, see Supplementary Fig. 9d.
Notably, the K+-loaded structure features densities in the S1, S3, and S4 positions of the SF of KdpA, although the S4 site was previously suggested to be lost in KdpFABC12 (Supplementary Fig. 8c). The nonprotein density in the S1 site is absent in the Rb+-loaded structure, probably because the mutation KdpAG232D changes the SF geometry (Supplementary Fig. 9c). The loss of a coordination site in the SF might also explain the reduced affinity of KdpFAG232DBC for K+ compared to wild-type KdpFABC10.
When comparing the individual nonprotein densities in both structures low-pass filtered to the same resolution, the densities in the KdpA SF S3 site and the KdpB CBS are increased ca. two-fold in intensity in the presence of Rb+ (Fig. 3b–d), close to the expected factor of 2.524,25,26. This suggests that the densities at both the beginning and the end of the intersubunit tunnel indeed correspond to ions, not waters, and, consequently, that the tunnel constitutes the ion translocation pathway. The effect is not as evident for ions in the middle of the intersubunit tunnel (Supplementary Fig. 9d), probably due to more dynamic and nonspecific ion coordination at these sites compared to the KdpA SF and KdpB CBS. Alternatively, these densities could represent waters in the intersubunit tunnel, as proposed recently12, although waters are not typically seen in EM at the obtained resolutions29. An unequivocal assignment of ions would require anomalous dispersion data from an X-ray diffraction approach.
The viability of ion translocation through the intersubunit tunnel was further probed by atomistic MD simulations of K+-loaded KdpFABC. The data revealed that the ions rearranged slightly during a short 20 ns MD relaxation, but are otherwise well coordinated in the tunnel (Fig. 4a). The simulations also showed that, in addition to direct protein contacts, water molecules form solvation shells around the K+ ions in the tunnel (Supplementary Fig. 10a). An analysis of the coordination energy for K+ along the tunnel by Adaptive Poisson-Boltzmann Solver (APBS) analysis was used as an approximate framework to compare ion positions in the tunnel, and indicated a favorable energy profile along the entire length, with notable energy minima in the KdpA SF, where K+ is fully dehydrated by the protein, and, interestingly, at the end of the intersubunit tunnel, just before the KdpB CBS (Fig. 4b, Supplementary Fig. 10b, c). Notably, this new ion position was taken up immediately, where K+ is primarily coordinated by KdpBD583. In addition, KdpBF232 aids ion coordination, either by direct cation–π stacking or by CH–π interaction with KdpBL262, which in turn coordinates the ion with its backbone carbonyl (Fig. 4c, Supplementary Fig. 10d). Since this position is located on the proximal side of KdpBF232 with respect to the CBS, we termed it the proximal binding site (PBS). When the five ions closest to the CBS were deleted, simulations revealed a rapid progression of two ions from KdpA towards the CBS (Video 1). The first ion moves quickly past the constriction formed by KdpBF232 to the described energy minimum, coordinated by KdpBD583 and the proximal phenylalanine face (PBS, Fig. 4c). Subsequently, the second ion moves along the intersubunit tunnel in a similar fashion, until being coordinated by cation–π stacking at the face of KdpBF232 distal to the CBS (distal binding site, DBS, Fig. 4d). These simulations not only support the translocation of ions through the intersubunit tunnel, but also suggest that ion propagation is supported by a pull-on mechanism mediated by the KdpBD583 energy well in the PBS at the end of the tunnel.
a Comparison of K+ positions from cryo-EM (gray) and at the endpoint of a short atomistic MD relaxation (purple). The ions rearrange slightly but remain in the intersubunit tunnel, indicating its viability for translocation. A new ion position is taken up adjacent to the CBS. b APBS coordination energy profile for K+ along the length of the intersubunit tunnel. Coordination is optimal at the KdpA SF, where ions are fully dehydrated by the protein, and immediately before the KdpB CBS, where they are bound by KdpBD583. c, d New coordination sites around KdpBF232 observed in atomistic MD simulations. During the progression towards the CBS, ions are likely coordinated by cation–π interactions on both faces of KdpBF232. Coordination in the PBS (c) is strong, with coordination supported by KdpBD583. Coordination in the DBS (d) is transient.
Moreover, the ion positions taken up in the MD simulations suggest that cation–π interactions with KdpBF232 play an important role in the progression of ions towards the CBS. Since the coordination in the PBS is energetically similar to that observed in the KdpA SF, an external force is likely required to push the ion onward into the CBS for transport, as is the case in the SF. The MD simulations used a point-charge force field, thus this process could not be fully resolved30. However, we postulate that the second ion found in the MD simulations, coordinated at the DBS, repels the first ion from the PBS, pushing it forward towards the CBS for subsequent transport. This could occur either through perturbation of the polarity of the phenylalanine π-electron clouds or simply through charge–charge repulsion, comparable to the proposed Coulomb knockon in the SFs of K+ channels31,32.
Three residues mediate ion progression and ATPase coupling
To further elucidate the proposed role of KdpBF232 in ion translocation, we mutated the residue to an isoleucine, which has a bulky side chain that fills the tunnel similarly to KdpBF232, but is unable to coordinate K+ via cation–π stacking. The KdpFABF232IC variant showed a similar K+-coupled ATPase activity as the wild type, but with an ~80% reduced transport rate (Fig. 5a–c). Atomistic MD simulations showed that the mutation does not greatly affect ion progression towards KdpB, attracted by the KdpBD583 energy well, and PBS and CBS can be populated. However, the DBS adjacent to KdpBF232I is lost, and ion binding at this site is no longer observed (Fig. 5d). The loss of ion coordination, together with the reduced transport rate, supports our hypothesis that the DBS next to KdpBF232 is required for the efficient forwarding of a K+ ion from the PBS to the CBS.
a ATPase activity of KdpFABC variants in the absence and presence of K+ and the P-type ATPase inhibitor orthovanadate (oV), normalized to K+-stimulated activity of wild-type (WT) KdpFABC. KdpFABF232IC retains K+-stimulated ATPase activity. The K+ mimic KdpBL228R uncouples ATP hydrolysis from K+, and increases resistance to orthovanadate. KdpBF232A uncouples ATP hydrolysis from K+ and largely increases the activity, while maintaining orthovanadate inhibition. Data points represent the average and error bars the standard deviation of measurements from technical replicates (–KCl | +KCl | +KCl +50 µM oV | +KCl +500 µM oV: WT n = 10 | 11 | 9 | 0, F232I n = 3 | 3 | 3 | 0, L228R n = 4 | 4 | 3 | 3, F232A n = 4 | 4 | 3 | 0) from at minimum biological triplicates (WT n = 9, F232I n = 3, L228R n = 3, F232A n = 3). b, c Transport activity of liposome-reconstituted KdpFABC variants. Decreasing ACMA fluorescence indicates K+ transport. Initial slopes were fitted linearly to obtain transport rates. Contrasting their ATPase activity, transport by KdpFABF232IC and KdpFABF232AC is slower than that of WT (reduced by 80 and 65%, respectively). KdpFABL228RC showed no transport. Data points represent the average and error bars the standard deviation of measurements from technical replicates (WT n = 26, F232I n = 9, F232A n = 23) from at minimum biological triplicates (WT n = 8, F232I = 3, F232A n = 3). d Computed densities of K+ (mesh) in atomistic MD simulations of KdpFABC variants. In WT, ions can occupy three main sites: the DBS and PBS at the KdpA/KdpB interface before and behind the constricting residue KdpBF232, and the CBS in KdpB. In KdpFABF232IC, ions can reach the PBS, coordinated by KdpBD583, but the DBS is lost. No significant ion passage past KdpBF232 was observed in KdpFABD583AC and KdpFABL228RC. Removing the steric hindrance in KdpFABF232AC once again allows ion passage to the PBS, but the DBS remains lost. Ion progression into the CBS in these simulations is possible because KdpBK586 is set as uncharged.
The fact that the ATPase activity is undisturbed in KdpFABF232IC further suggests that cation binding to the PBS is sufficient to trigger ATP hydrolysis. We postulate that the neutralization of the negative charge of KdpBD583 by cation binding could be the stimulus, as variant KdpFABD583AC has previously been shown to uncouple ATP hydrolysis from K+ and abolish K+ transport33. The lack of transport is likely based on the loss of the energy well at the end of the intersubunit tunnel, as MD simulations with KdpFABD583AC showed that ions are no longer effectively pulled past the constriction formed by KdpBF232 (Fig. 5d). To elucidate whether neutralization at the PBS is in fact responsible for stimulating ATP hydrolysis, we placed a positively charged side chain into the PBS, mimicking K+ binding. The activity of KdpFABL228RC fully mirrored that of KdpFABD583AC: the ATPase activity level was similar to WT and uncoupled from K+, K+ transport was abolished, and in MD simulations no ions passed KdpBF232 (Fig. 5a, b, d).
In addition to its role in ion coordination at the PBS and forwarding of K+ to the CBS, KdpBF232 appears to present a significant steric constriction in the intersubunit tunnel, preventing ion transition to the PBS unless pulled by the negatively charged KdpBD583 (Fig. 5d). To probe this, we investigated the impact of the KdpBF232A mutation where, unlike KdpBF232I, not only the possibility of cation–π interactions, but also steric hindrance is abolished. As seen for KdpBF232I, ions can reach the CBS and PBS but do not occupy the DBS in the MD simulations (Fig. 5d). Moreover, simulations on the double mutant KdpBF232A/D583A showed that ions can reach the CBS even in the absence of the KdpBD583A energy well when the steric hindrance is removed (Supplementary Fig. 10e). Functional assays of KdpFABF232AC, like KdpFABF232IC, revealed a significantly reduced transport rate (Fig. 5b, c), supporting the importance of cation-π interactions and the DBS for efficient ion progression towards the CBS. Unexpectedly however, KdpFABF232AC showed a K+-uncoupled and enhanced ATPase activity (Fig. 5a). The uncoupling suggests that ATP hydrolysis is stimulated by a K+-independent neutralization of the PBS, as described above for KdpFABD583AC and KdpFABL228RC. A possible explanation could be an unspecific protonation of KdpBD583 via a continuous water wire along the intersubunit tunnel, which is otherwise sterically blocked in the WT and KdpFABF232IC. Thus, in addition to being an element in ion coordination and progression, KdpBF232 may also fulfill a gatekeeper role in KdpFABC by preventing unspecific stimulation of ATP hydrolysis by K+-independent neutralization of KdpBD583.
KdpBF232 modulates the rate-limiting step in KdpFABC
In addition to uncoupling ATP hydrolysis from K+, KdpFABF232AC also shows a three- to four-fold increase in Vmax of ATP hydrolysis when compared to stimulated wild-type KdpFABC (Fig. 5a, Supplementary Fig. 11). By contrast, the Km value for ATP is similar. Interestingly, the uncoupled variants KdpFABL228RC and KdpFABD583AC33 showed a Vmax comparable to that of stimulated wild-type KdpFABC. This discrepancy between the uncoupled variants suggests that ATPase uncoupling and deregulation are separate processes. The KdpFABD583AC variant was previously shown to have a decreased sensitivity to orthovanadate, which was interpreted as indicative of an accelerated E2/E1 transition, as orthovanadate binds to the protein in the post-hydrolysis E2 state and stalls turnover33. Similarly, the KdpBL228R mutation led to an increased resistance to inhibition by orthovanadate (Fig. 5a). The proposed accelerated E2/E1 transition could be explained by the lack of ions in the CBS in these variants, which otherwise have to be displaced before the return to the E1 state. By contrast, KdpBF232A does not increase the resistance to orthovanadate, suggesting that the mutation must affect the catalytic cycle differently. We propose that the increase in ATPase activity observed for KdpFABF232AC is based on the acceleration of the rate-limiting step in KdpFABC turnover, the E1-P/E2-P transition34, which is characterized by large rearrangements of the A domain12,17. KdpBF232 lies in TM3, which is directly connected to the A domain (Supplementary Fig. 12a). Therefore, the plasticity of the A domain could be modulated by the large phenylalanine side chain, regulating the E1-P/E2-P transition. Thus, in addition to its involvement in ion coordination and gatekeeping the CBS, KdpBF232 may modulate the movement of the KdpB-A domain, in part regulating the rate-limiting step of KdpFABC turnover.
Discussion
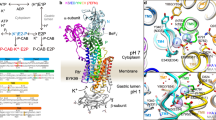
By combining structural, biochemical, and MD simulation data, we demonstrate how K+ is translocated through KdpFABC. Beyond confirming the previously proposed intersubunit tunnel translocation model, our data allow us to propose a molecular mechanism for ion propagation and ATPase coupling, particularly involving residues KdpBF232, KdpBD583, and KdpBK586 (Fig. 6). Notably, the here proposed model also explains previously reported functional effects of mutations in and around the CBS (for details see Supplementary Fig. 13).
a In the E1 ground state (Early E1), KdpBK586 is protonated and KdpBD583 deprotonated, forming the energy well at the end of the intersubunit tunnel. b ATP binds and a first ion is rapidly pulled past the constriction of KdpBF232 into the PBS, coordinated by KdpBD583 and assisted by cation–π stacking with KdpBF232 (E1·ATP transitory 1). c When a second ion reaches the DBS, π cloud polarization is perturbed, slightly repelling the first ion forward towards the CBS (E1·ATP transitory 2). d Upon a protonation switch between KdpBD583/K586, the PBS ion moves forward into the CBS, with coordination supported by KdpBK586 (Late E1/E1-P). The energy well in the PBS is lost by protonation of KdpBD583, preventing the ion from being pulled back or a second ion from being pulled forward from the DBS. e Upon E1-P/E2-P transition, the protonation switches back to its ground state, with a conformational shift of the now positively charged KdpBK586 forcing the ion out of the CBS (5—E2-P/E2). In the E2/E1 transition, KdpBK586 acts as a built-in counterion.
In the E1 state, a first K+ ion is drawn past the constriction formed by KdpBF232 to the energy minimum at the PBS, generated largely by the negatively charged KdpBD583 (Fig. 6a). Coordination is aided in part by direct cation–π stacking to KdpBF232, or by KdpBL262, which in turn is CH–π stacked to KdpBF232. The arrival of a second ion from the tunnel at the DBS perturbs the polarization of the KdpBF232 π-system and electrostatically repels the ion in the PBS towards the CBS (Fig. 6b). We propose that this induces a protonation switch between KdpBD583 and KdpBK586, neutralizing both (Fig. 6c). This hypothesis is supported by their predicted pKa values close to physiological pH (pKa of KdpBK586: 7.5; pKa of KdpBD583: 7.2), suggesting that protonation swapping is energetically inexpensive. The proton switching consequently allows the ion in the PBS to move forward into the CBS without being pulled back by the energy well (Fig. 6d). Coordination in the CBS is facilitated by deprotonated KdpBK586, as observed in previous E1 structures of KdpFABC12,16. This forwarding of a K+ ion by a protonation switch mechanism is in good agreement with ion behavior in MD simulations: in the doubly charged state, the ion binds strongly at the PBS; when the protonation states are switched (doubly neutral state), the ion is repelled forward into the CBS once a second ion approaches the DBS (Supplementary Fig. 14a, b).
During the post-hydrolysis E1-P/E2-P transition, the protonation states of KdpBD583/K586 likely revert back to their initial states, such that the now positively charged lysine side chain displaces the K+ from the CBS, as previously proposed17 (Fig. 6e). A lipid at the KdpA/KdpB interface might support the E1-P/E2-P transition, as it was suggested to stabilize the E2 conformation12. We assign the lipid as CL (Fig. 2, Supplementary Fig. 7), and the accelerated turnover in the presence of CL probably reflects the effect on the rate-limiting step. Similar lipid-mediated effects have been shown for other P-type ATPases35,36.
Recent structural observations in the E2 state suggest that KdpBK586 pushes the ion into an adjacent low-affinity site, from where it is released through a previously observed inward-open half-channel into the cytosol12,17. We tested this hypothesis using MD simulations, and saw that ions were released from the CBS in the E2 state via the proposed half-channel, but did not enter this low-affinity release site (Supplementary Fig. 14c). Moreover, ions placed in this site rapidly moved back towards the CBS, as shown by K+ distances to KdpBK586 in the CBS (Supplementary Fig. 14d). From there, they followed the same exit pathway as before, indicating that the proposed low-affinity site likely does not represent a K+ binding site. In accordance with these observations, the physiological necessity for such a low-affinity site remains unclear, as ion release appears to be possible without traversing it.
The duration of ion release from the CBS is variable, taking up to 250 ns in simulations (Supplementary Fig. 14e). This may explain the accelerated E2/E1 transition in KdpFABD583AC and KdpFABL228RC, in which ions do not reach the CBS and no longer need to be displaced. During the E2/E1 transition, KdpBK586 acts as a built-in counterion, as previously suggested17 (Fig. 6e). Alternating access is granted by a constriction at the KdpA/KdpB interface in the E2 state, which prevents new ions from entering the CBS before the return to the E1 conformation (Supplementary Fig. 14f).
But at what point in the proposed transport cycle does ATP hydrolysis occur? Our data suggest that hydrolysis is induced by the binding of K+ to the PBS, which neutralizes KdpBD583 and leaves KdpBK586 at the CBS positively charged (Fig. 6b, c). This trigger is possibly mediated through TM5 of KdpB, harboring KdpBD583 and KdpBK586, which directly connects to the P domain (Supplementary Fig. 12b). Hence, to allow K+ transport to the cytoplasm, the forwarding of the ion from the PBS to the CBS must be faster than the transition from E1·ATP to the inward-open E2-P. This scenario is plausible, as the E1-P/E2-P transition is the rate-limiting step of KdpFABC34, regulated by residue KdpBF232.
In fact, our data place a particular importance on residue KdpBF232. It appears to have a three-fold role in KdpFABC turnover: (1) it acts as a gatekeeper to prevent unspecific access to the PBS and CBS from the intersubunit tunnel, thereby preventing an uncoupling of ATP hydrolysis from K+ ; (2) it is involved in ion coordination and progression via its π-electron system; and (3) it regulates the rate-limiting step of KdpFABC, the E1-P/E2-P transition. As such, KdpBF232 directly links ion propagation and turnover. The elaborate coupling system may explain the low transport rate of KdpFABC compared to other P-type ATPases and the strict coupling of transport to ATP hydrolysis37. Accordingly, KdpBF232, like KdpBD583/K58633, is highly conserved among KdpB sequences from different species (Supplementary Fig. 15).
Notably, all elements involved in ion progression and coupling, individually or in combinations, have also been proposed for other P-type ATPases. Cation–π coordination by phenylalanine residues has been implicated in ion transport by the Na+/K+-ATPase and in the sarcoplasmic Ca2+ pump SERCA. In the Na+/K+-ATPase (P2C family), a phenylalanine is involved in the dehydration of ions entering the CBS38. Like KdpBF232, this residue lies in the TM helix connected to the A domain, and its mutation to alanine resulted in an acceleration of the E1-P/E2-P transition39,40. In SERCA (P2A family), a structurally equivalent phenylalanine residue was suggested to be involved in stabilizing the E2 conformation, favoring the E1-P/E2-P transition41. In the Zn2+ ATPase ZntA (P1B family), metal binding and ATPase coupling are facilitated by an aspartate/lysine pair in the CBS, while a conserved tyrosine residue controls access to the CBS42,43. The lysine also acts as a built-in counterion in the E2 state, enabling electrogenic transport without a counterion, as in KdpFABC. A similar mechanism was shown for the H+ ATPase (P3A family), in which the CBS is formed by an arginine and an aspartate, while an asparagine acts as an H+ gatekeeper, replacing the phenylalanine/tyrosine in KdpFABC and ZntA, respectively44,45. This indicates that metal ATPases require the aromatic residue for interaction with the ion. The H+/K+ ATPase (P2C family) features a lysine in the CBS that is suggested to expel the primary ion in the E2 state before counterion binding46, in part reflecting the function proposed for KdpBK586. Prevention of counterion binding was also observed upon mutation of an arginine into the corresponding position of the Na+/K+ ATPase47. The fact that different elements identified in various P-type ATPase families are all present in KdpB suggests that it may be a close descendant of a common ancestor of cation pumps.
In light of these similarities, it remains elusive why KdpFABC includes an additional channel-like subunit in the transport process. The SF of KdpA is likely crucial to the particularly high substrate affinity and specificity. Due to its low sequence conservation to the classical TVGYG motif of tetrameric K+ channels, SF discrimination in KdpA was suggested to base on a different mechanism12,16. Functional data already showed an absolute selectivity of KdpFABC for K+, and, contrary to most K+ channels, wild-type KdpFABC even excludes Rb+. Specific mutations in the SF only allow the passage of Rb+ and NH4+, and under no circumstances is Na+ permitted9,10. A question not addressed here is how K+ ions so selectively pass the SF to reach the intersubunit tunnel. Ion permeation in K+ channels with the canonical TVGYG SFs is mediated by a Coulomb knock-on31,32. Despite the significantly altered selectivity filter, we observe the occupancy of three coordination sites (S1, S3, S4) in the SF, and similarly optimal coordination moieties for K+ binding in the S2 site. While cryo-EM data do not allow to precisely quantify the occupancies of bound ligands, the presence of densities at three SF-binding sites, which are stably and simultaneously occupied in our MD simulations, strongly suggests a mechanism for ion entry into KdpFABC similar to that found in tetrameric K+ channels. If so, this might explain the high selectivity of KdpFABC when compared to other SKT members, which showed fewer coordination sites48,49. Compared to canonical K+ channels and the other SKT members, KdpFABC has a very low Km for K+ transport of 2 µM8. The low Km, essential for the function of KdpFABC as a high-affinity K+ transporter, could be enabled by the energy minimum at the PBS in KdpB, which may augment the energetics of the SF knock-on to grant efficient ion progression. Furthermore, KdpC could promote high-affinity K+ binding, as its sequence is related to β-subunits of other P-type ATPases, which are involved in substrate binding7,50.
The data presented here verifies that ion transport in KdpFABC occurs via the unique intersubunit tunnel through KdpA and KdpB, and allows us to describe how ATP hydrolysis and K+ transport are coupled in the chimeric complex. Moreover, we propose an intricate molecular system for the forwarding of ions into the CBS for transport, with key elements conserved in other P-type ATPases.
Methods
Cloning and protein production
Escherichia coli kdpFABC was brought into expression vector pBXC3H using FX cloning51. Point mutations based on this construct were generated by site-directed mutagenesis, creating different variants listed in Table 1.
KdpFABC variants were produced in E. coli LB2003 cells (available from the Hänelt group upon request) transformed with plasmids encoding the relevant variant and grown in 6 or 12 l KML with 100 µg/ml ampicillin. Cultures were inoculated to OD600 0.1, induced with 0.002% L-arabinose at OD600 1.2, and harvested one hour after induction. For functional studies, KdpFABC variants were produced, purified, and characterized in parallel with wild-type KdpFABC to ensure comparability of measurements.
KdpFABC purification
Purification of KdpFABC variants for structural studies was performed as previously described for wild-type KdpFABC17. Purification of KdpFABC variants for functional studies was performed identically, omitting the final size exclusion chromatography step after anion exchange chromatography.
Cryo-EM sample and grid preparation
For the cryo-EM sample resulting in the 3.1 Å structure in the E1·ATP state with K+, KdpFABD307NC was used. The mutation KdpBD307N prevents ATP hydrolysis and stalls the complex in the E1 state52. Purified protein was concentrated to 3.1 mg/ml, and supplemented with 50 mM KCl and 5 mM AMPPCP to stabilize the complex in the E1·ATP conformation prior to grid preparation.
For the cryo-EM sample resulting in the 3.2 Å structure in the E1·ATP state with Rb+, KdpFAG232DBS162AC was used. The mutation KdpAG232D in the KdpA SF allows the passage of Rb+9,10, while KdpBS162 is the site of an inhibitory phosphorylation that inhibits turnover16,21. Purified protein was concentrated to 4 mg/ml, and supplemented with 100 mM RbCl and 1 mM ATP before incubation for 5 min at room temperature to displace any remaining K+ in the complex with Rb+. Turnover was stopped by the addition of 10 mM AMPPCP to stabilize the complex in the E1·ATP conformation prior to grid preparation.
Holey-carbon cryo-EM grids (Quantifoil Au R1.2/1.3, 300 mesh) were twice glow-discharged at 15 mA for 45 s. 2.8 µl of KdpFABC sample were applied to grids, blotted for 2–6 s in a VitroBot (Mark IV, ThermoFisher) at 4 °C and 100% humidity, and subsequently plunge-frozen in liquid ethane and stored in liquid nitrogen.
Cryo-EM data collection
Cryo-EM data were automatically collected using SerialEM software53,54 (Thermo Fisher Scientific) on a 200 keV Talos Arctica microscope (Thermo Fisher Scientific) equipped with a post-column energy filter (Gatan) in zero-loss mode with a 20 eV slit and a 100 µM aperture with a K2 Summit detector (Gatan). Images were recorded at a pixel size of 1.012 Å (calibrated magnification of 49407×), a defocus range from −0.2 to −2.0 μm, an exposure time of 9 s, a subframe exposure time of 150 ms (60 frames), and a total electron exposure on the specimen level of about 52 electrons per Å2. Data collection was optimized by restricting acquisition to regions displaying optimal sample thickness using an in-house written script55, and the data quality monitored by on-the-fly processing using FOCUS software56.
Cryo-EM data processing
The SBGrid57 software package tool was used to manage the software packages.
K+-loaded KdpFABD307NC
A total of 5,831 dose-fractionated cryo-EM images were recorded and subjected to motion-correction and dose-weighting of frames by MotionCor258. The CTF parameters were estimated on the movie frames by ctffind4.1.1359. Bad images showing contamination, a defocus below −0.5 or above −2.0 μm, or a bad CTF estimation were discarded, resulting in 3,530 images used for further analysis with the software package RELION 3.0.860. First, crYOLO 1.3.161 was used to automatically pick 331,673 particles using a loose threshold. Particle coordinates were imported in RELION 3.0.860, and the particles were extracted with a box size of 240 pixels. False positives or particles belonging to low-abundance classes were removed in several rounds of 2D classification, resulting in 249,092 particles. For 3D classification and refinement, the map of the previously generated E1 conformation EMD-025717 was used as reference for the first round, and the best output class was used in subsequent jobs in an iterative way. Particles belonging to the best classes were selected, resulting in 160,776 particles. Sequentially, several rounds of CTF refinement60 were performed, using per-particle CTF estimation. In the last refinement iteration, a mask excluding the micelle was used and the refinement was continued until convergence (focused refinement), yielding a final map for the E1·ATP state at a resolution of 3.5 Å, and 3.1 Å after post-processing and masking, sharpened using an isotropic b-factor of −92 Å2. No symmetry was imposed during 3D classification or refinement.
Rb+-loaded KdpFAG232DBS162AC
A total of 22,046 dose-fractionated cryo-EM images were recorded and subjected to motion-correction and dose-weighting of frames by MotionCor258. The CTF parameters were estimated on the movie frames by ctffind4.1.1359. Bad images showing contamination, a defocus below −0.5 or above −2.0 μm or a bad CTF estimation were discarded, resulting in 14,947 images used for further analysis with the software package RELION 3.1.060. First, crYOLO 1.3.161 was used to automatically pick 756,834 particles using a loose threshold. Particle coordinates were imported in RELION 3.1.060, and the particles were extracted with a box size of 240 pixels. False positives or particles belonging to low-abundance classes were removed in several rounds of 2D classification, resulting in 469,824 particles. For 3D classification and refinement, the map of the previously generated E1 conformation EMD-025717 was used as reference for the first round, and the best output class was used in subsequent jobs in an iterative way. Particles belonging to the best class were selected, resulting in 276,980 particles. Sequentially, several rounds of CTF refinement60 were performed, using per-particle CTF estimation, before subjecting the dataset to a round of focused 3D classification with no image alignment, using a mask on the flexible AN domains of KdpB62, which resulted in a cleaned dataset of 196,682 particles. This dataset was subjected to another round of CTF refinement. In the last refinement iteration, a mask excluding the micelle was used and the refinement was continued until convergence (focused refinement), yielding a final map at a resolution of 3.6 Å before masking and 3.2 Å after masking, sharpened using an isotropic b-factor of −130 Å2. No symmetry was imposed during 3D classification or refinement.
For both datasets, local resolution estimates were calculated by RELION. All resolutions were estimated using the 0.143 cutoff criterion63 with gold-standard Fourier shell correlation (FSC) between two independently refined half maps. During post-processing, the approach of high-resolution noise substitution was used to correct for convolution effects of real-space masking on the FSC curve64.
Model building and validation
The cryo-EM structure of KdpFABC in an E1 conformation 6HRA17 was split into the membrane domain: KdpA, KdpB (residues 1–88, 216–274, and 570–682,), KdpC, and KdpF, and the three cytosolic domains of KdpB: KdpB-P (residues 275–314 and 451–569), KdpB-N (residues 315–450) and KdpB-A (residues 89–215). Initially, all fragments were docked into the obtained cryo-EM map using UCSF Chimera65. The connections between the four KdpB segments were modeled manually in Coot66. The initial model was then subjected to an iterative process of real-space refinement using Phenix.real_space_refinement with secondary structure restraints67,68, followed by manual inspection and adjustments in Coot66. Rubidium ions, cardiolipins, and AMPPCP were modeled into the cryo-EM maps. The final model was refined in real space with Phenix.real_space_refinement with secondary structure restraints67,68. For validation of the refinement, FSC (FSCsum) between the refined model and the final map was determined. To monitor the effects of potential over-fitting, random shifts (up to 0.5 Å) were introduced into the coordinates of the final model, followed by refinement against the first unfiltered half-map. The FSC between this shaken-refined model and the first half-map used during validation refinement is termed FSCwork, and the FSC against the second half-map, which was not used at any point during refinement, is termed FSCfree. The marginal gap between the curves describing FSCwork and FSCfree indicate no over-fitting of the model. The geometries of the atomic model was evaluated using MolProbity69.
Tunnel calculations
Pore calculations through the selectivity filter of the channel-like subunit KdpA were performed using the software HOLE70. For calculations, the PDB of KdpA, without cofactors or other subunits and aligned with the translocation pore parallel to the z-axis, was probed using a Conolly probe of radius 0.9 Å.
Radius calculations of the intersubunit tunnel were performed using CAVER Analyst71. Calculations were performed on PDB models without ions using a probe radius of 0.9 Å to allow estimation of constrictions in the tunnel. These settings result in a set of unspecific tunnels, from which the relevant tunnel was selected via comparison with previously calculated tunnels in 6HRA17. The intersubunit tunnel between KdpA and KdpB was visualized using HOLLOW with a probe radius of 1.1 Å and a starting point immediately below the KdpA SF72.
Ion density analysis
Identified densities in the intersubunit tunnel of the K+-loaded and the Rb+-loaded E1·ATP map presented here were low-pass filtered to a resolution of 3.2 Å. For these filtered maps, the ion densities in the CBS, the intersubunit tunnel, and SF were quantified in Coot66, by determining the minimum sigma for these densities to be visualized. For global comparison of all possible ion densities, these values were then normalized against the value obtained for the S3 position in the K+-loaded map for each applied filter. For comparisons of individual densities, values were normalized to the value obtained for the specific ion in the K+-loaded map.
The expected scattering ratio of Rb/K was estimated as follow. A simplified estimation is obtained by using the formula Zˆ4/3, which is independent of the electron voltage24,25,26. With Z = 19 for K and 37 for Rb, the Rb/K ratio is 2.43. A better approximation, valid for elements with Z < ~100 and taking into account the relativistic speed of the electrons, which makes the ratio dependent on the acceleration voltage, is described in Reimer and Kohl25, formula 5.41. Using beta = v/c = 0.7 for 200 kV gives a Rb/K ratio of 2.59 (v is the speed of the electrons, c is the speed of light). Based on these calculations, using an expected factor of 2.5 seems appropriate.
ATPase assay
ATP hydrolysis by purified KdpFABC variants was observed by malachite green ATPase assay73. In brief, 2 mM ATP, 1 mM KCl, and indicated lipids or inhibitors were mixed to the respective conditions. Reactions were started by the addition of 0.5–1.0 µg protein and carried out for 5 min at 37 °C. Lipid stocks were prepared as previously described36.
Growth complementation assay
The growth complementation assays were performed as previously described74. In brief, E. coli LB2003, a strain lacking all endogenous K+ uptake systems, was transformed with plasmids encoding His-tagged KdpFABC variants. LB2003 transformed with empty vector pBAD18 or plasmid pBXC3H-KdpFABD307NC, encoding for an inactive variant, served as negative controls. Growth was monitored for 24 h at different K+ concentrations (1–115 mM, referred to as K1–K115). At K10 and below, the strain only grows sufficiently if the produced protein complements the lacking K+ transport systems. Protein production was confirmed by SDS-PAGE and subsequent western blotting analysis of a K30 sample after 24 h using an anti-His antibody from mouse (dilution 1:3000, Sigma–Aldrich, cat.no. H1029) and secondary anti-mouse IgG-HRP antibody produced in goat (dilution 1:20,000, Sigma–Aldrich, cat.no. A2554).
Reconstitution and ACMA-based liposome transport assay
Liposomes were prepared from E. coli polar lipid extract. Five milligrams of dry lipids were resuspended to 10 mg/ml in hot (55 °C) reconstitution buffer (15 mM Tris-HCl pH 7.5, 2 mM MgCl2, 50 mM KCl), homogenized by sonication and subjected to three freeze-thaw cycles. Liposomes were diluted to a concentration of 4 mg/ml in reconstitution buffer, and extruded to a diameter of 400 nm. Extruded liposomes were destabilized by titration of 10% Triton-X-100 to saturation. Detergent-solubilized protein was added to destabilized liposomes at a mass ratio of 1:5 and incubated at room temperature for 30 min. Detergent was removed by the gradual addition of Bio-BeadsTM SM-2 in 40 mg/ml wet weight intervals, with incubation times between additions as follows: 15 min at room temperature, 15 min at 4 °C, 30 min at 4 °C, 1 h at 4 °C, and overnight at 4 °C. In the final detergent removal step, 60 mg/ml Bio-Beads were added and incubated for 1 h at 4 °C. The supernatant was extracted, proteoliposomes pelleted by ultracentrifugation for 30 min at 80,000 × g, washed once in reconstitution buffer and pelleted again by ultracentrifugation for 30 min at 80,000 × g. Proteoliposomes were resuspended to a final concentration of 5 mg/ml in reconstitution buffer for further experiments.
K+ translocation by reconstituted KdpFABC was tested by a modified ACMA-based flux assay75. Proteoliposomes were diluted to 1 mg/ml and supplemented with 2 μM ACMA (Ex. 410 nm, Em. 480 nm) and 1 μM CCCP. Transport was initialized by the addition of 0.2 mM ATP, and ended by the addition of 0.2 µM valinomycin. Transport rates were obtained by linear fitting of the initial 50 s of transport.
Molecular dynamics simulations
CGMD simulations
CG systems were built using the deposited coordinates for PDBs 5MRW, 6HRA, 6HRB, 7BGY, and 7NNL. Protein atoms were converted to the CG Martini force field76,77, using the Martini 3.0.b.3.2 open beta78. Additional bonds of 500 kJ mol−1nm−2 were applied between all protein backbone beads within 1 nm. Proteins were built into membranes composed of 10% CL, 23% POPG, and 67% POPE, using the insane protocol79. CL parameters were used as described in Corey et al., 202128. All systems were solvated with Martini waters and Na+ and Cl− ions to a neutral charge and 150 mM. Systems were minimized using the steepest descents method, followed by 1 ns equilibration with 5 fs time steps, then by 100 ns equilibration with 20 fs time steps, before 5 × 4 µs production simulations using 20 fs time steps, all in the NPT ensemble with the V-rescale thermostat and semi-isotropic Parrinello-Rahman pressure coupling80,81.
Protein-lipid contact analyses were run on the final 3 µs of each trajectory, where contact was set at an inter-residue minimum distance of 0.6 nm, and the fraction of frames for which contact occurs plotted.
Atomistic MD simulations
For simulations of KdpFABC, the structural model of 7NNL was truncated by removing the KdpB soluble domains in the regions KdpBV100-G196 and KdpBD302-L558. Positional restraints (1000 kJ mol−1 nm−2) were applied to backbone atoms C, Cα, and N to prevent shifts in the input structure. Alternatively, simulations were built using the full-length KdpFABC in the E2 state 7BGY. Protein atoms were described using the CHARMM36 force field, and inserted into a 67% POPE, 23% POPG, 10% CL lipid bilayer solvated with TIP3P water and 150 mM K+/Cl− using CHARMM-GUI82,83,84. For the majority of the simulations, the protonation states of all side chains were set to default apart from KdpBK586, and KdpAE370, which had pKas of 7.5, and 9.1, respectively, based on analysis with PropKa3.185. Additional systems were built in which KdpBD583 and KdpBK586 were either both charged or both neutral, as well as a neutral KdpBE370 (Supplementary Fig. 14a, b).
Systems were energy minimized using the steepest descents method, and subsequently equilibrated with positional restraints on heavy atoms for 100 ps in the NPT ensemble at 310 K with the V-rescale thermostat and semi-isotropic Parrinello-Rahman pressure coupling80,81. For both stages, positional restraints of 1000 kJ mol−1 nm−2 were applied to the K+ in the tunnel. Production simulations were run in triplicate with 2 fs time steps for ca. 250 ns unless specified.
Ion density analyses were run using the VolMap tool of VMD86 with the default settings, taken over 3 × 250 ns of simulation for each system.
All atomistic and CGMD simulations were run in Gromacs 201987.
APBS calculations
Adaptive Poisson-Boltzmann Solver (APBS) calculations88 were performed for K+ along the intersubunit tunnel. The first set of calculations (Fig. 3b) were for K+ positions taken from a 20 ns MD relaxation simulation. Note that positions from multiple MD snapshots were taken to provide maximum coverage of the tunnel (see Supplementary Fig. 10b for positions). Alternatively, the input structural protein and ion coordinates from 7NNL were used, following steepest descents energy minimization (Supplementary Fig. 10c).
For each K+ position, a system was built-in CHARMM36 comprising KdpFABC and the single K+ ion at that position. The coordinates were then relaxed using steepest descents energy minimization, and processed using PDB2PQR89, setting KdpAK586 and KdpBE370 to neutral states. APBS values for K+ were determined, assuming an ionic radius of 2.172 Å. The default settings for APBS were used as produced by the APBS webserver (https://server.poissonboltzmann.org/).
Sequence alignment
Conservation of sequences in KdpB across different species was evaluated by sequence comparison using Clustal Omega90, with sequences from previously published comparisons16 to ensure species diversity.
Figure preparation
Figures were prepared using UCSF Chimera65, UCSF ChimeraX91, PyMOL, VMD86, OriginPro 2016, and GraphPad Prism 8.
Data availability
The cryo-EM map and model of K+-loaded KdpFABD307NC E1·ATP were deposited in the EMDB and wwPDB with accession codes EMD-12478 and 7NNL, respectively. The cryo-EM map and model of Rb+-loaded KdpFAG232DBS162AC E1·ATP were deposited in the EMDB and wwPDB with accession codes EMD-12482 and 7NNP, respectively. Source data are provided with this paper for all functional data; the minimum dataset is provided in a source data file. Further data supporting the findings in this manuscript are available upon reasonable request. Source data are provided with this paper.
References
Epstein, W., Buurman, E., McLaggan, D. & Naprstek, J. Multiple mechanisms, roles and controls of K+ transport in Escherichia coli. Biochem. Soc. Trans. 21, 1006–1010 (1993).
Altendorf, K. et al. Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. Suppl. 643, 137–146 (1998).
Stautz, J. et al. Molecular Mechanisms for Bacterial Potassium Homeostasis. J. Mol. Biol. 433, 166968 (2021).
Durell, S. R., Bakker, E. P. & Guy, H. R. Does the KdpA subunit from the high affinity K+-translocating P-type Kdp-ATPase have a structure similar to that of K+ channels? Biophys. J. 78, 188–199 (2000).
Hesse, J. E. et al. Sequence homology between two membrane transport ATPases, the Kdp-ATPase of Escherichia coli and the Ca2+-ATPase of sarcoplasmic reticulum. Proc. Natl Acad. Sci. USA 81, 4746–4750 (1984).
Gaßel, M., Möllenkamp, T., Puppe, W. & Altendorf, K. The KdpF subunit is part of the K+-translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 274, 37901–37907 (1999).
Shull, G. E., Lane, L. K. & Lingrel, J. B. Amino-acid sequence of the β-subunit of the (Na+ + K+)ATPase deduced from a cDNA. Nature 321, 429–431 (1986).
Rhoads, D. B., Waters, F. B. & Epstein, W. Cation transport in Escherichia coli VIII. potassium transport mutants. J. Gen. Physiol. 67, 325–341 (1976).
Schrader, M. et al. Replacement of glycine 232 by aspartic acid in the KdpA subunit broadens the ion specificity of the K+-translocating KdpFABC complex. Biophys. J. 79, 802–813 (2000).
van der Laan, M., Gassel, M. & Altendorf, K. Characterization of amino acid substitutions in KdpA, the K+-binding and -translocating subunit of the KdpFABC complex of Escherichia coli. J. Bacteriol. 184, 5491–5494 (2002).
Bertrand, J., Altendorf, K. & Bramkamp, M. Amino acid substitutions in putative selectivity filter regions III and IV in KdpA alter ion selectivity of the KdpFABC complex from Escherichia coli. J. Bacteriol. 186, 5519–5522 (2004).
Sweet, M. E. et al. Structural basis for potassium transport by KdpFABC. Proc. Natl Acad. Sci. USA 118, e2105195118 (2021).
Diskowski, M., Mikusevic, V., Stock, C. & Hänelt, I. Functional diversity of the superfamily of K+ transporters to meet various requirements. Biol. Chem. 396, 1003–1014 (2015).
Albers, R. W. Biochemical aspects of active transport. Annu. Rev. Biochem. 36, 727–756 (1967).
Post, R. L., Hegyvary, C. & Kume, S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 (1972).
Huang, C.-S., Panyella Pedersen, B. & Stokes, D. L. Crystal structure of the potassium-importing KdpFABC membrane complex. Nature 546, 681–685 (2017).
Stock, C. et al. Cryo-EM structures of KdpFABC suggest a K+ transport mechanism via two inter-subunit half-channels. Nat. Commun. 9, 4971 (2018).
Serrano, R. & Portillo, F. Catalytic and regulatory sites of yeast plasma membrane H+-ATPase studied by directed mutagenesis. Biochim. Biophys. Acta 1018, 195–199 (1990).
Toustrup-Jensen, M. & Vilsen, B. Importance of conserved Thr214 in domain A of the Na+,K+-ATPase for stabilization of the phosphoryl transition state complex in E2P dephosphorylation. J. Biol. Chem. 278, 11402–11410 (2003).
Anthonisen, A. N., Clausen, J. D. & Andersen, J. P. Mutational analysis of the conserved TGES loop of sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 281, 31572–31582 (2006).
Sweet, M. E. et al. Serine phosphorylation regulates the P-type potassium pump KdpFABC. eLife 9, e55480 (2020).
Kühlbrandt, W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 (2004).
Greie, J. C. & Altendorf, K. The K+-translocating KdpFABC complex from Escherichia coli: a P-type ATPase with unique features. J. Bioenerg. Biomembr. 39, 397–402 (2007).
Egerton, R. F. Measurement of inelastic/elastic scattering ratio for fast electrons and its use in the study of radiation damage. Phys. Status Solidi 37, 663–668 (1976).
Reimer, L. & Kohl, H. Scattering and phase contrast. Springe. Ser. Opt. Sci. 36, 193–269 (2008).
Franken, L. E., Grünewald, K., Boekema, E. J. & Stuart, M. C. A. A Technical introduction to transmission electron microscopy for soft-matter: imaging, possibilities, choices, and technical developments. Small 16, 1906198 (2020).
Jensen, A. M. L., Sørensen, T. L. M., Olesen, C., Møller, J. V. & Nissen, P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 25, 2305–2314 (2006).
Corey, R. A. et al. Identification and assessment of cardiolipin interactions with E. coli inner membrane proteins. bioRxiv https://doi.org/10.1101/2021.03.19.436130 (2021).
Vonck, J. & Mills, D. J. Advances in high-resolution cryo-EM of oligomeric enzymes. Curr. Opin. Struct. Biol. 46, 48–54 (2017).
Lin, F. Y. & MacKerell, A. D. Improved modeling of cation-π and anion-ring interactions using the drude polarizable empirical force field for proteins. J. Comput. Chem. 41, 439–448 (2020).
Kopec, W. et al. Direct knock-on of desolvated ions governs strict ion selectivity in K+ channels. Nat. Chem. 10, 813–820 (2018).
Öster, C. et al. The conduction pathway of potassium channels is water free under physiological conditions. Sci. Adv. 5, eaaw6756 (2019).
Bramkamp, M. & Altendorf, K. Single amino acid substitution in the putative transmembrane helix V in KdpB of the KdpFABC complex of Escherichia coli uncouples ATPase activity and ion transport. Biochemistry 44, 8260–8266 (2005).
Damnjanovic, B. & Apell, H. J. Role of protons in the pump cycle of KdpFABC investigated by time-resolved kinetic experiments. Biochemistry 53, 3218–3228 (2014).
Hossain, K. R. & Clarke, R. J. General and specific interactions of the phospholipid bilayer with P-type ATPases. Biophys. Rev. 11, 353–364 (2019).
Subramani, S., Perdreau-Dahl, H. & Morth, J. P. The magnesium transporter A is activated by cardiolipin and is highly sensitive to free magnesium in vitro. eLife 5, e11407 (2016).
Pedersen, B. P., Stokes, D. L. & Apell, H.-J. The KdpFABC complex—K+ transport against all odds. Mol. Membr. Biol. 35, 1–34 (2019).
Morth, J. P. et al. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat. Rev. Mol. Cell Biol. 12, 60–70 (2011).
Vilsen, B. & Toustrup-Jensen, M. Importance of transmembrane segment M3 of Na+,K+-ATPase for control of conformational changes and the cytoplasmic entry pathway for Na+. Ann. N. Y. Acad. Sci. 986, 50–57 (2003).
Toustrup-Jensen, M. & Vilsen, B. Functional consequences of alterations to Ile279, Ile 283, Glu284, His285, Phe286, and His288 in the NH2-terminal part of transmembrane helix M3 of the Na+,K+-ATPase. J. Biol. Chem. 278, 38653–38664 (2003).
Laursen, M. et al. Cyclopiazonic acid is complexed to a divalent metal ion when bound to the sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 284, 13513–13518 (2009).
Okkeri, J. & Haltia, T. The metal-binding sites of the zinc-transporting P-type ATPase of Escherichia coli. Lys693 and Asp714 in the seventh and eighth transmembrane segments of ZntA contribute to the coupling of metal binding and ATPase activity. Biochim. Biophys. Acta 1757, 1485–1495 (2006).
Wang, K. et al. Structure and mechanism of Zn2+-transporting P-type ATPases. Nature 514, 518–522 (2014).
Pedersen, B. P., Buch-Pedersen, M. J., Preben Morth, J., Palmgren, M. G. & Nissen, P. Crystal structure of the plasma membrane proton pump. Nature 450, 1111–1114 (2007).
Ekberg, K., Wielandt, A. G., Buch-Pedersen, M. J. & Palmgren, M. G. A conserved asparagine in a P-type proton pump is required for efficient gating of protons. J. Biol. Chem. 288, 9610–9618 (2013).
Munson, K., Garcia, R. & Sachs, G. Inhibitor and ion binding sites on the gastric H,K-ATPase. Biochemistry 44, 5267–5284 (2005).
Holm, R. et al. Arginine substitution of a cysteine in transmembrane helix M8 converts Na+,K+-ATPase to an electroneutral pump similar to H+,K+-ATPase. Proc. Natl Acad. Sci. USA 114, 316–321 (2017).
Vieira-Pires, R. S., Szollosi, A. & Morais-Cabral, J. H. The structure of the KtrAB potassium transporter. Nature 496, 323–328 (2013).
Cao, Y. et al. Crystal structure of a potassium ion transporter, TrkH. Nature 471, 336–340 (2011).
Axelsen, K. B. & Palmgren, M. G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 (1998).
Geertsma, E. R. & Dutzler, R. A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry 50, 3272–3278 (2011).
Puppe, W., Siebers, A. & Altendorf, K. The phosphorylation site of the Kdp-ATPase of Escherichia coli: site-directed mutagenesis of the aspartic acid residues 300 and 307 of the KdpB subunit. Mol. Microbiol. 6, 3511–3520 (1992).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Schorb, M., Haberbosch, I., Hagen, W. J. H., Schwab, Y. & Mastronarde, D. N. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019).
Rheinberger, J., Oostergetel, G., Resch, G. P. & Paulino, C. Optimized cryo-EM data-acquisition workflow by sample-thickness determination. Acta Crystallogr. Sect. D. Struct. Biol. 77, 565–571 (2021).
Biyani, N. et al. Focus: The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Zheng, S. Q. et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Hiraizumi, M., Yamashita, K., Nishizawa, T. & Nureki, O. Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 365, 1149–1155 (2019).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D. Struct. Biol. 75, 861–877 (2019).
Afonine, P. V., Headd, J. J., Terwilliger, T. C. & Adams, P. D. New Tool: phenix.real_space_refine. Comput. Crystallogr. Newsl. 4, 43–44 (2013).
Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Jurcik, A. et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 34, 3586–3588 (2018).
Ho, B. K. & Gruswitz, F. HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008).
Carter, S. G. & Karl, D. W. Inorganic phosphate assay with malachite green: an improvement and evaluation. J. Biochem. Biophys. Methods 7, 7–13 (1982).
Tholema, N. et al. All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J. Biol. Chem. 280, 41146–41154 (2005).
Mikušević, V. et al. A channel profile report of the unusual K+ channel KtrB. J. Gen. Physiol. 151, 1357–1368 (2019).
Marrink, S. J., Risselada, H. J., Yefimov, S., Tieleman, D. P. & De Vries, A. H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007).
Monticelli, L. et al. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 4, 819–834 (2008).
Souza, P. C. T. et al. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat. Methods 18, 382–388 (2021).
Wassenaar, T. A., Ingólfsson, H. I., Böckmann, R. A., Tieleman, D. P. & Marrink, S. J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 11, 2144–2155 (2015).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Jo, S., Kim, T. & Im, W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2, e880 (2007).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Olsson, M. H. M., Søndergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Humphrey, W., Dalke, A. & Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Berendsen, H. J. C., van der Spoel, D. & van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995).
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001).
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We thank Werner Kühlbrandt for access to the vitrobot and Sonja Welsch and Janina Stautz for freezing the cryo-EM grids. J.M.S. thanks Paul JN Böhm for assistance with cloning, cell growth, and purification. C.P. thanks Jan Rheinberger for EM support and Michiel Punter for IT support. P.J.S. acknowledges the University of Warwick Scientific Computing Research Technology Platform for computational access. The work was funded by the NWO Veni grant (722.017.001) and the NWO Start-Up grant (740.018.016) to C.P. as well as by the DFG Emmy Noether grant (HA6322/3-1), and the Life Science Bridge Award by the Aventis Foundation to IH. Research in P.J.S.’s lab is funded by Wellcome (208361/Z/17/Z), the MRC (MR/S009213/1) and BBSRC (BB/P01948X/1, BB/R002517/1, and BB/S003339/1). J.M.S. is funded by the State of Hesse in the LOEWE Schwerpunkt TRABITA and R.A.C. is funded by Wellcome (208361/Z/17/Z). This project made use of time on ARCHER, ARCHER2, and JADE granted via the UK High-End Computing Consortium for Biomolecular Simulation, HECBioSim (http://hecbiosim.ac.uk), supported by EPSRC (grant no. EP/R029407/1).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
J.M.S.: Validation, formal analysis, investigation, writing—original draft, writing—review and editing, visualization. R.A.C.: Validation, formal analysis, investigation, writing—original draft, writing—review and editing, visualization. L.H.: Validation, formal analysis, investigation, writing—original draft, writing—review and editing, visualization. C.S.: Validation, formal analysis, investigation, writing—original draft, writing—review and editing, visualization. P.J.S.: Conceptualization, writing—review and editing, supervision, project administration, funding acquisition. C.P.: Conceptualization, writing—review and editing, supervision, project administration, funding acquisition. I.H.: Conceptualization, writing—review and editing, supervision, project administration, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewers for their contributions to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41467_2021_25242_MOESM3_ESM.mov
Supplementary Movie 1: Progression of K<sup>+</sup> through the intersubunit tunnel of KdpFABC in atomistic MD simulations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Silberberg, J.M., Corey, R.A., Hielkema, L. et al. Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC. Nat Commun 12, 5098 (2021). https://doi.org/10.1038/s41467-021-25242-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-021-25242-x
This article is cited by
-
KdpD is a tandem serine histidine kinase that controls K+ pump KdpFABC transcriptionally and post-translationally
Nature Communications (2024)
-
Tuning phenylalanine fluorination to assess aromatic contributions to protein function and stability in cells
Nature Communications (2023)
-
LipIDens: simulation assisted interpretation of lipid densities in cryo-EM structures of membrane proteins
Nature Communications (2023)
-
Wie Bakterien die Aufnahme von Kaliumionen regulieren
BIOspektrum (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.